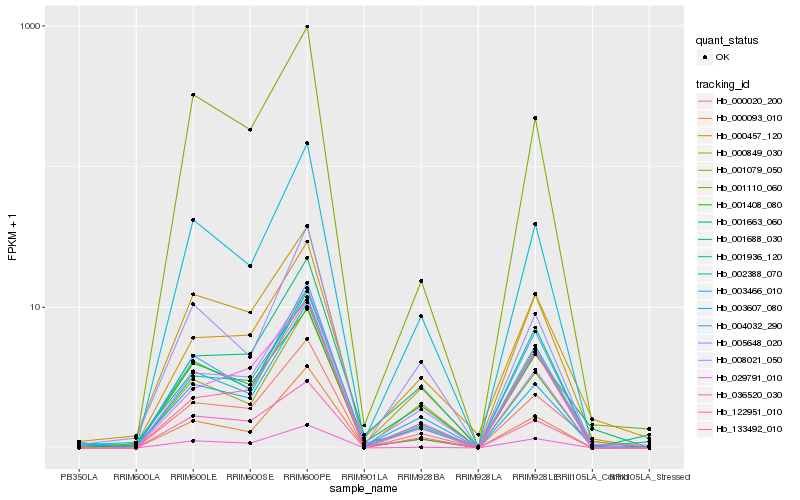
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000020_200 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_B00411 [Eucalyptus grandis] |
| 2 |
Hb_001663_060 |
0.0807387329 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 3 |
Hb_000849_030 |
0.0951874417 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 4 |
Hb_003466_010 |
0.1008512096 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 5 |
Hb_008021_050 |
0.1021085765 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_036520_030 |
0.1082763256 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 7 |
Hb_000457_120 |
0.1097457586 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 8 |
Hb_029791_010 |
0.1116350502 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 9 |
Hb_003607_080 |
0.1134214152 |
- |
- |
PREDICTED: putative kinase-like protein TMKL1 [Jatropha curcas] |
| 10 |
Hb_001079_050 |
0.1158620441 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 11 |
Hb_001936_120 |
0.1164307853 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 12 |
Hb_122951_010 |
0.11886101 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 13 |
Hb_000093_010 |
0.1221754055 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 14 |
Hb_133492_010 |
0.1256038786 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_001688_030 |
0.1271235881 |
- |
- |
hypothetical protein JCGZ_10716 [Jatropha curcas] |
| 16 |
Hb_004032_290 |
0.1277721813 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_001110_060 |
0.128588467 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 18 |
Hb_002388_070 |
0.1310394794 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_001408_080 |
0.1317535289 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 20 |
Hb_005648_020 |
0.1332751456 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |