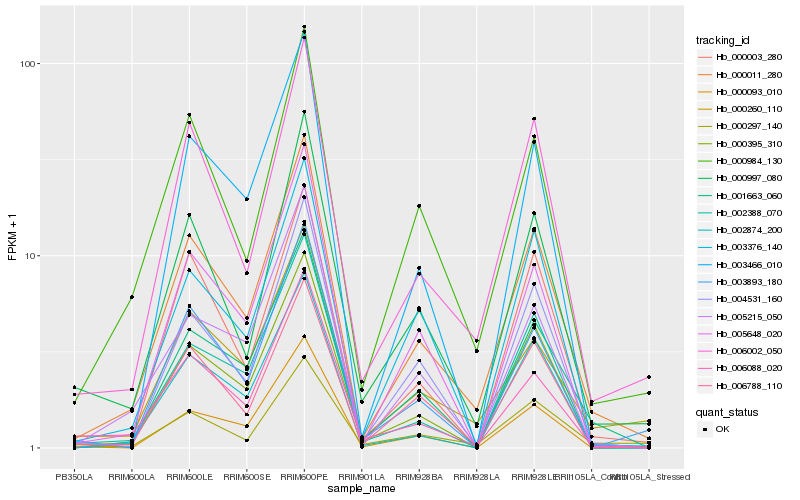
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_280 |
0.0 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 2 |
Hb_006002_050 |
0.0866486882 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 3 |
Hb_000984_130 |
0.0960886116 |
- |
- |
tubulin [Ornithogalum longebracteatum] |
| 4 |
Hb_001663_060 |
0.1036833291 |
- |
- |
ribonuclease t2, putative [Ricinus communis] |
| 5 |
Hb_000395_310 |
0.1059518312 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000997_080 |
0.1069460988 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006788_110 |
0.1118657917 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 8 |
Hb_005648_020 |
0.1126970461 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 9 |
Hb_000260_110 |
0.1151924243 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 10 |
Hb_000003_280 |
0.1156925272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004531_160 |
0.1157458398 |
- |
- |
PREDICTED: transcription factor bHLH68-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_006088_020 |
0.1181594566 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 13 |
Hb_003376_140 |
0.1184754144 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 14 |
Hb_000297_140 |
0.1198234234 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 15 |
Hb_003466_010 |
0.120326259 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 16 |
Hb_002388_070 |
0.1207718885 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_005215_050 |
0.1228711903 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 18 |
Hb_003893_180 |
0.1228829958 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 19 |
Hb_002874_200 |
0.1233107746 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 20 |
Hb_000093_010 |
0.1253059173 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |