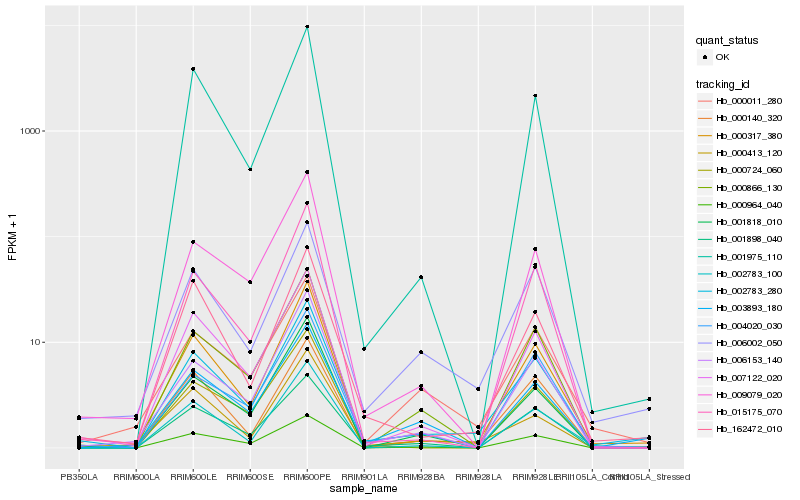
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002783_280 |
0.0 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 2 |
Hb_000317_380 |
0.0898459683 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 3 |
Hb_006002_050 |
0.1015415791 |
- |
- |
PREDICTED: uncharacterized protein LOC105638083 [Jatropha curcas] |
| 4 |
Hb_007122_020 |
0.1127578432 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 5 |
Hb_000140_320 |
0.1184440192 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 6 |
Hb_162472_010 |
0.1188749536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000413_120 |
0.1234459071 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 8 |
Hb_001898_040 |
0.1247230394 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 9 |
Hb_004020_030 |
0.1250471405 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 10 |
Hb_000866_130 |
0.1253263769 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |
| 11 |
Hb_000724_060 |
0.1254001222 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 12 |
Hb_003893_180 |
0.1266158697 |
- |
- |
PREDICTED: uncharacterized protein LOC105648771 [Jatropha curcas] |
| 13 |
Hb_006153_140 |
0.1272350205 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 14 |
Hb_000011_280 |
0.1277657122 |
- |
- |
pectinesterase family protein [Populus trichocarpa] |
| 15 |
Hb_000964_040 |
0.1280688081 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |
| 16 |
Hb_002783_100 |
0.1280800142 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 17 |
Hb_001818_010 |
0.1283038394 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 18 |
Hb_009079_020 |
0.128566498 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 19 |
Hb_015175_070 |
0.1288645851 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 20 |
Hb_001975_110 |
0.1297440831 |
- |
- |
- |