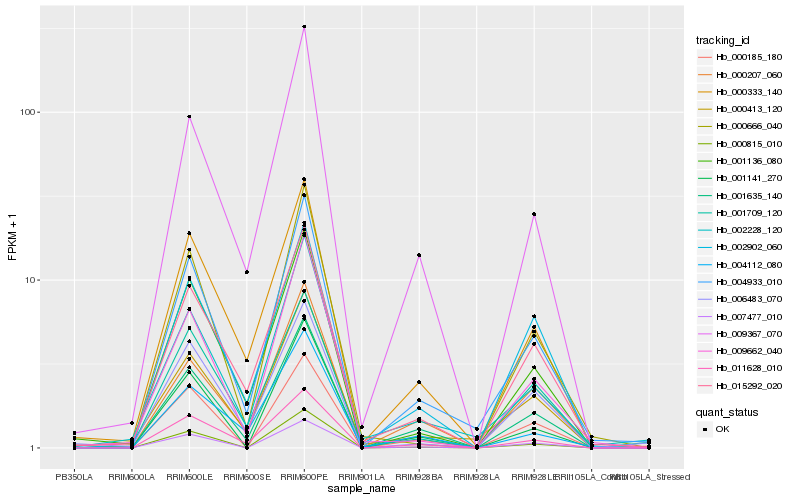
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004933_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 2 |
Hb_001635_140 |
0.092145774 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_011628_010 |
0.0937735167 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 4 |
Hb_002228_120 |
0.0948947206 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 5 |
Hb_000413_120 |
0.1005946977 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 6 |
Hb_000333_140 |
0.1056643983 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 7 |
Hb_015292_020 |
0.1068873442 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_007477_010 |
0.1086820704 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA9 [Phoenix dactylifera] |
| 9 |
Hb_001136_080 |
0.1133945278 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 10 |
Hb_009367_070 |
0.1143979181 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 11 |
Hb_001709_120 |
0.1171844954 |
- |
- |
hypothetical protein RCOM_1313640 [Ricinus communis] |
| 12 |
Hb_000185_180 |
0.1173960398 |
- |
- |
hypothetical protein CISIN_1g037968mg, partial [Citrus sinensis] |
| 13 |
Hb_000666_040 |
0.1180124624 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 14 |
Hb_000815_010 |
0.118726234 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105117338 [Populus euphratica] |
| 15 |
Hb_009662_040 |
0.1221281686 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 16 |
Hb_001141_270 |
0.1229726673 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 17 |
Hb_000207_060 |
0.1242967951 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 18 |
Hb_002902_060 |
0.1244780664 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 19 |
Hb_004112_080 |
0.1267070921 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 20 |
Hb_006483_070 |
0.1267841015 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |