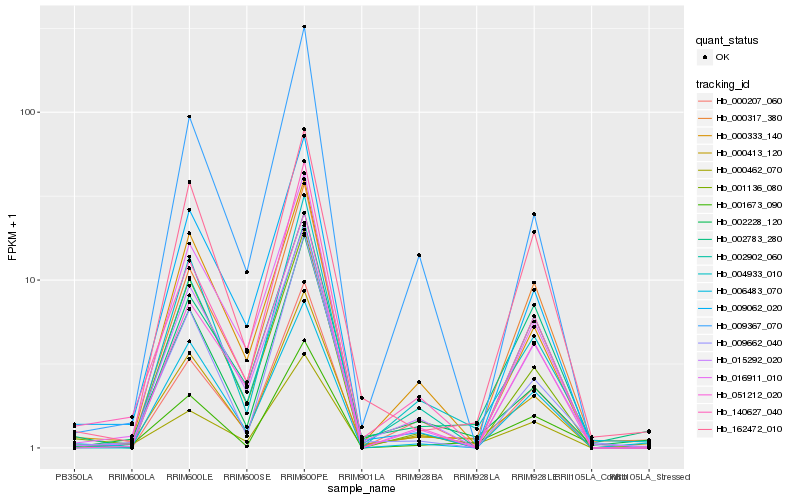
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000413_120 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 2 |
Hb_002228_120 |
0.0817391336 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 3 |
Hb_004933_010 |
0.1005946977 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 4 |
Hb_015292_020 |
0.1008297741 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_140627_040 |
0.1059771525 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 6 |
Hb_000333_140 |
0.1131417714 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 7 |
Hb_009062_020 |
0.1169993925 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 8 |
Hb_162472_010 |
0.1197678125 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001673_090 |
0.1222888657 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |
| 10 |
Hb_016911_010 |
0.1224817831 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 11 |
Hb_001136_080 |
0.1226789378 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 12 |
Hb_009662_040 |
0.1229119601 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 13 |
Hb_002783_280 |
0.1234459071 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 14 |
Hb_006483_070 |
0.1255575778 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_009367_070 |
0.126391524 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 16 |
Hb_051212_020 |
0.1267392846 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 17 |
Hb_000462_070 |
0.128338591 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 18 |
Hb_000207_060 |
0.1289059484 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 19 |
Hb_000317_380 |
0.129149496 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 20 |
Hb_002902_060 |
0.1291856885 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |