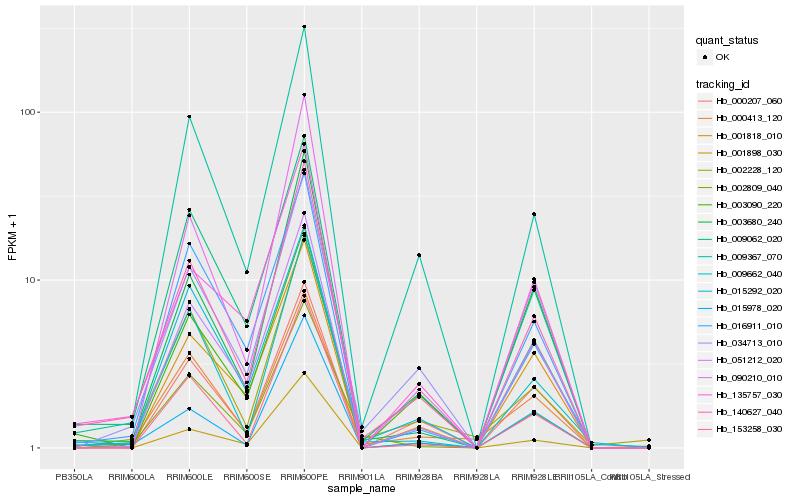
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_140627_040 |
0.0 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 2 |
Hb_034713_010 |
0.0815844285 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |
| 3 |
Hb_003680_240 |
0.0877815733 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 4 |
Hb_009367_070 |
0.088040341 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 5 |
Hb_051212_020 |
0.0959375054 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 6 |
Hb_000207_060 |
0.1014081809 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 7 |
Hb_009062_020 |
0.1028788785 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 8 |
Hb_009662_040 |
0.1039438556 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 9 |
Hb_135757_030 |
0.1050153502 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 10 |
Hb_000413_120 |
0.1059771525 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 11 |
Hb_090210_010 |
0.1072745686 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 12 |
Hb_001818_010 |
0.1081829065 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 13 |
Hb_153258_030 |
0.1117893499 |
- |
- |
hypothetical protein RCOM_0853150 [Ricinus communis] |
| 14 |
Hb_016911_010 |
0.1127393846 |
- |
- |
hypothetical protein POPTR_0010s15010g [Populus trichocarpa] |
| 15 |
Hb_002809_040 |
0.1130466715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002228_120 |
0.113664836 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 17 |
Hb_003090_220 |
0.1149464368 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 18 |
Hb_015978_020 |
0.115155062 |
- |
- |
Multidrug resistance protein ABC transporter family protein, putative [Theobroma cacao] |
| 19 |
Hb_015292_020 |
0.1187540732 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_001898_030 |
0.1193606748 |
- |
- |
protein kinase-coding resistance protein [Nicotiana repanda] |