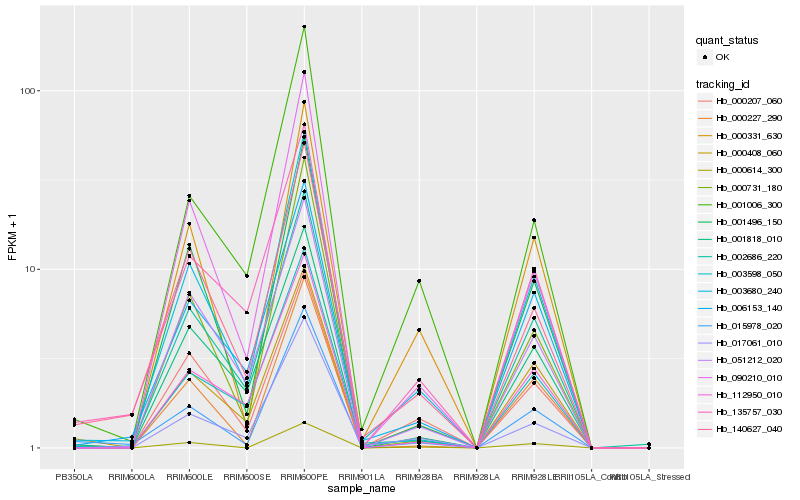
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003680_240 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 2 |
Hb_002686_220 |
0.0831797171 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 3 |
Hb_003598_050 |
0.084030073 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 4 |
Hb_006153_140 |
0.0870220518 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 5 |
Hb_000331_630 |
0.0872770589 |
- |
- |
hypothetical protein B456_008G203900 [Gossypium raimondii] |
| 6 |
Hb_140627_040 |
0.0877815733 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 7 |
Hb_017061_010 |
0.0883546109 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 5-like [Jatropha curcas] |
| 8 |
Hb_001818_010 |
0.0911315548 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 9 |
Hb_015978_020 |
0.0938686654 |
- |
- |
Multidrug resistance protein ABC transporter family protein, putative [Theobroma cacao] |
| 10 |
Hb_000408_060 |
0.0955522405 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 11 |
Hb_090210_010 |
0.0965041969 |
- |
- |
neutral alpha-glucosidase ab precursor, putative [Ricinus communis] |
| 12 |
Hb_001496_150 |
0.096806118 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 13 |
Hb_051212_020 |
0.0981834955 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 14 |
Hb_000227_290 |
0.0997710611 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 15 |
Hb_000207_060 |
0.0999701864 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 16 |
Hb_112950_010 |
0.1003012229 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 17 |
Hb_001006_300 |
0.100431318 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 18 |
Hb_135757_030 |
0.1012138522 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 19 |
Hb_000731_180 |
0.1019142717 |
- |
- |
hypothetical protein POPTR_0004s01820g [Populus trichocarpa] |
| 20 |
Hb_000614_300 |
0.1026018492 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Jatropha curcas] |