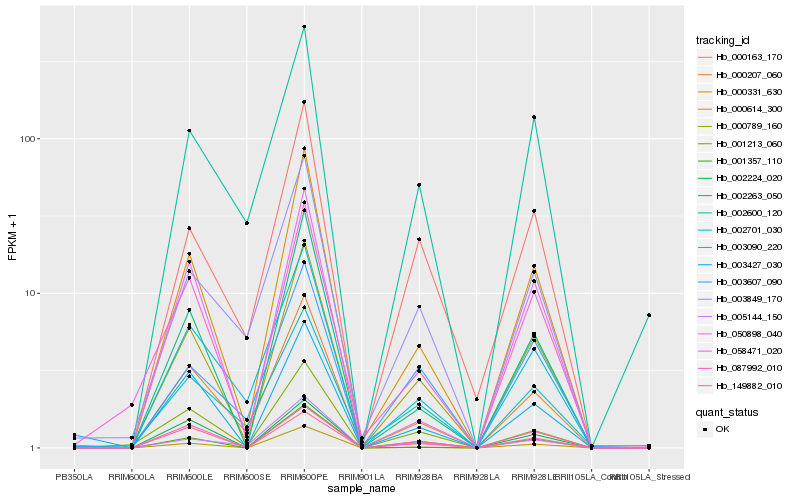
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000789_160 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 2 |
Hb_000331_630 |
0.0688165598 |
- |
- |
hypothetical protein B456_008G203900 [Gossypium raimondii] |
| 3 |
Hb_000614_300 |
0.0976877897 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Jatropha curcas] |
| 4 |
Hb_003427_030 |
0.0988549905 |
- |
- |
- |
| 5 |
Hb_000207_060 |
0.1037287685 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 6 |
Hb_002263_050 |
0.1099119151 |
- |
- |
hypothetical protein POPTR_0016s06160g [Populus trichocarpa] |
| 7 |
Hb_000163_170 |
0.1132616666 |
- |
- |
- |
| 8 |
Hb_058471_020 |
0.1146749002 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 9 |
Hb_001213_060 |
0.1218937792 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 10 |
Hb_002600_120 |
0.1284209252 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 11 |
Hb_002224_020 |
0.1293554525 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 12 |
Hb_087992_010 |
0.1296972484 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_003090_220 |
0.1308773246 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 14 |
Hb_003607_090 |
0.1332377276 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 15 |
Hb_149882_010 |
0.1344547459 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Populus euphratica] |
| 16 |
Hb_003849_170 |
0.1353482547 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 17 |
Hb_050898_040 |
0.1359035301 |
- |
- |
- |
| 18 |
Hb_001357_110 |
0.1378116079 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 19 |
Hb_005144_150 |
0.1386555227 |
- |
- |
hypothetical protein VITISV_013572 [Vitis vinifera] |
| 20 |
Hb_002701_030 |
0.1394540308 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |