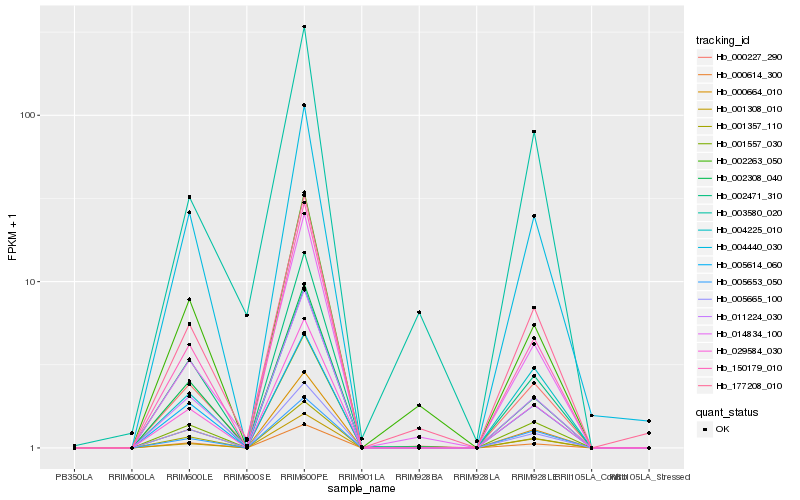
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_177208_010 |
0.0 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor, partial [Jatropha sp. CONN 198700232] |
| 2 |
Hb_000227_290 |
0.0675109968 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 3 |
Hb_001357_110 |
0.0775905134 |
- |
- |
endo-1,4-beta-glucanase, putative [Ricinus communis] |
| 4 |
Hb_014834_100 |
0.0840519082 |
- |
- |
PREDICTED: probable aquaporin TIP-type [Gossypium raimondii] |
| 5 |
Hb_029584_030 |
0.0891434646 |
- |
- |
hypothetical protein JCGZ_07310 [Jatropha curcas] |
| 6 |
Hb_000664_010 |
0.0922554947 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 4-like [Jatropha curcas] |
| 7 |
Hb_011224_030 |
0.0942040782 |
- |
- |
PREDICTED: oligopeptide transporter 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000614_300 |
0.0965615149 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Jatropha curcas] |
| 9 |
Hb_004225_010 |
0.0982194251 |
- |
- |
- |
| 10 |
Hb_003580_020 |
0.0999391303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001308_010 |
0.1016410436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001557_030 |
0.1023501974 |
- |
- |
hypothetical protein CICLE_v10005306mg [Citrus clementina] |
| 13 |
Hb_005653_050 |
0.1025945276 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3-sialyltransferase [Jatropha curcas] |
| 14 |
Hb_004440_030 |
0.1035401482 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 15 |
Hb_002263_050 |
0.1038868833 |
- |
- |
hypothetical protein POPTR_0016s06160g [Populus trichocarpa] |
| 16 |
Hb_005665_100 |
0.1045568031 |
- |
- |
PREDICTED: RING-H2 finger protein ATL63-like [Populus euphratica] |
| 17 |
Hb_002471_310 |
0.1046623185 |
- |
- |
PREDICTED: vacuolar iron transporter 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002308_040 |
0.1056249245 |
- |
- |
methyladenine glycosylase family protein [Populus trichocarpa] |
| 19 |
Hb_005614_060 |
0.105971314 |
- |
- |
Uncharacterized protein TCM_019750 [Theobroma cacao] |
| 20 |
Hb_150179_010 |
0.1060616026 |
- |
- |
hypothetical protein JCGZ_21079 [Jatropha curcas] |