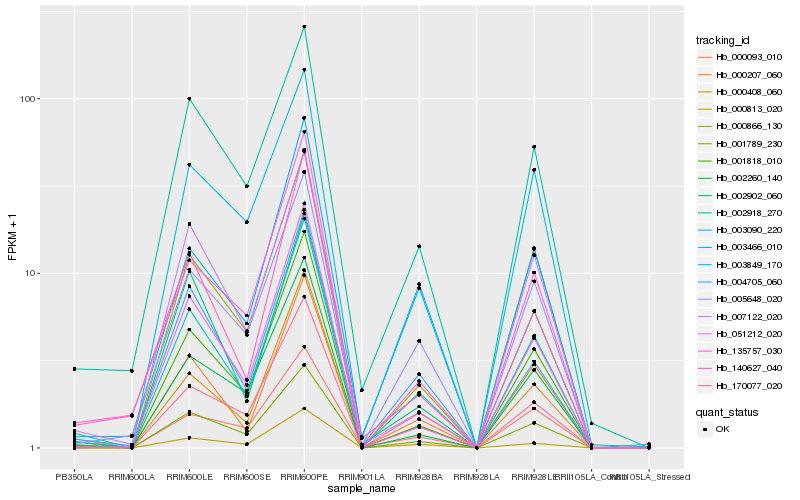
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003090_220 |
0.0 |
- |
- |
PREDICTED: copper transporter 4 [Jatropha curcas] |
| 2 |
Hb_000207_060 |
0.0485959953 |
- |
- |
PREDICTED: uncharacterized protein LOC105634368 [Jatropha curcas] |
| 3 |
Hb_001789_230 |
0.0649910263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001818_010 |
0.0767330642 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 5 |
Hb_000408_060 |
0.0775647989 |
- |
- |
hypothetical protein POPTR_0018s11360g [Populus trichocarpa] |
| 6 |
Hb_005648_020 |
0.0915796046 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM1 [Jatropha curcas] |
| 7 |
Hb_170077_020 |
0.0960772497 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 8 |
Hb_004705_060 |
0.0979428744 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 9 |
Hb_002260_140 |
0.0991267957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003849_170 |
0.1026504121 |
- |
- |
hypothetical protein JCGZ_17850 [Jatropha curcas] |
| 11 |
Hb_051212_020 |
0.1054690474 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 12 |
Hb_000866_130 |
0.10656012 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |
| 13 |
Hb_000813_020 |
0.1103564271 |
- |
- |
PREDICTED: metacaspase-3-like [Jatropha curcas] |
| 14 |
Hb_002918_270 |
0.1107949487 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 15 |
Hb_135757_030 |
0.1108689626 |
- |
- |
PREDICTED: actin-depolymerizing factor 5 [Jatropha curcas] |
| 16 |
Hb_000093_010 |
0.1126833565 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 17 |
Hb_003466_010 |
0.1134071394 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 18 |
Hb_002902_060 |
0.1135020841 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 19 |
Hb_140627_040 |
0.1149464368 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 20 |
Hb_007122_020 |
0.1152088339 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |