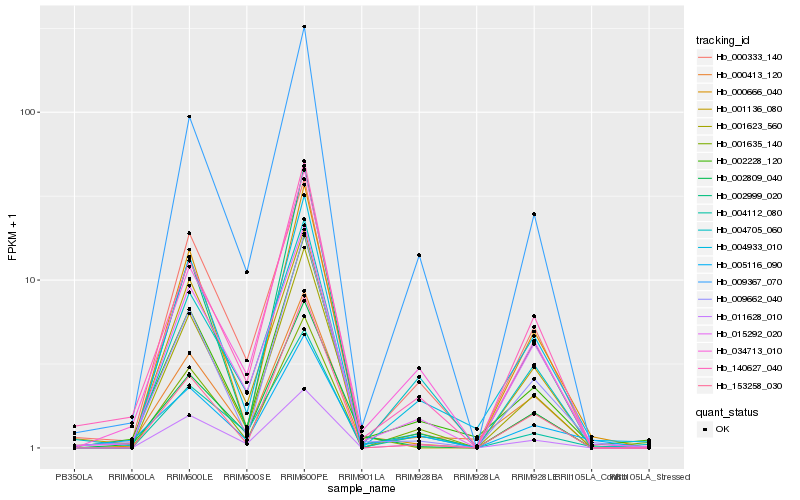
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002228_120 |
0.0 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 2 |
Hb_000413_120 |
0.0817391336 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 3 |
Hb_004933_010 |
0.0948947206 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 4 |
Hb_009662_040 |
0.1019691798 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 5 |
Hb_009367_070 |
0.1035518164 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 6 |
Hb_034713_010 |
0.1101008364 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |
| 7 |
Hb_140627_040 |
0.113664836 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 8 |
Hb_004112_080 |
0.1196638939 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 9 |
Hb_001635_140 |
0.1236545842 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000333_140 |
0.1249469924 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 11 |
Hb_015292_020 |
0.1260052165 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_002999_020 |
0.1294739051 |
- |
- |
PREDICTED: ribonuclease 3-like [Jatropha curcas] |
| 13 |
Hb_004705_060 |
0.1300642479 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 14 |
Hb_011628_010 |
0.1310826741 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 15 |
Hb_000666_040 |
0.1320356168 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 16 |
Hb_002809_040 |
0.1326051266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001623_560 |
0.1347146821 |
- |
- |
late embryogenesis abundant protein group 9 protein [Arachis hypogaea] |
| 18 |
Hb_153258_030 |
0.1354202062 |
- |
- |
hypothetical protein RCOM_0853150 [Ricinus communis] |
| 19 |
Hb_001136_080 |
0.1368461062 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 20 |
Hb_005116_090 |
0.1369095322 |
- |
- |
conserved hypothetical protein [Ricinus communis] |