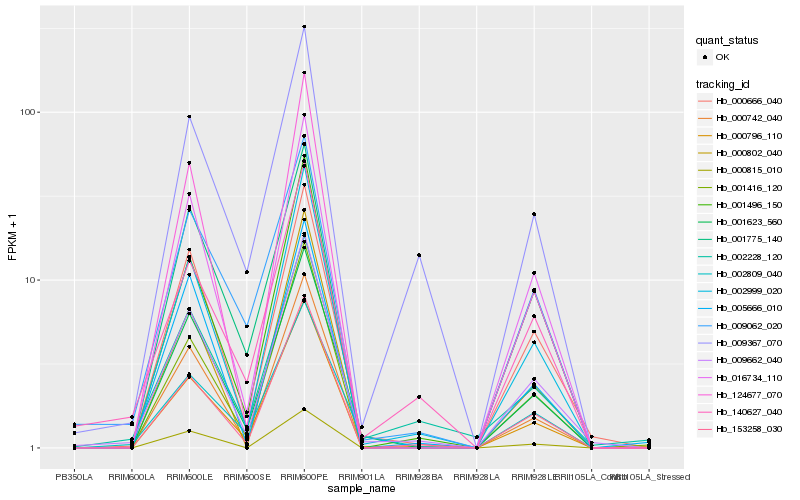
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009662_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 2 |
Hb_000666_040 |
0.0625318805 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 3 |
Hb_002999_020 |
0.0753261944 |
- |
- |
PREDICTED: ribonuclease 3-like [Jatropha curcas] |
| 4 |
Hb_153258_030 |
0.0872151817 |
- |
- |
hypothetical protein RCOM_0853150 [Ricinus communis] |
| 5 |
Hb_016734_110 |
0.0885806499 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 6 |
Hb_001623_560 |
0.0902388546 |
- |
- |
late embryogenesis abundant protein group 9 protein [Arachis hypogaea] |
| 7 |
Hb_000742_040 |
0.0918819615 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 8 |
Hb_000796_110 |
0.0949821962 |
- |
- |
PREDICTED: probable pectinesterase 53 [Populus euphratica] |
| 9 |
Hb_002228_120 |
0.1019691798 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 10 |
Hb_140627_040 |
0.1039438556 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 11 |
Hb_000815_010 |
0.1050948872 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105117338 [Populus euphratica] |
| 12 |
Hb_002809_040 |
0.1069026433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001496_150 |
0.1080198394 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 14 |
Hb_001416_120 |
0.1091604867 |
- |
- |
caspase, putative [Ricinus communis] |
| 15 |
Hb_009062_020 |
0.1117556213 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP20-2, chloroplastic [Cucumis melo] |
| 16 |
Hb_124677_070 |
0.1120656557 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 17 |
Hb_005666_010 |
0.1165738363 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 18 |
Hb_009367_070 |
0.1179369147 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 19 |
Hb_000802_040 |
0.1194338113 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |
| 20 |
Hb_001775_140 |
0.1197330516 |
- |
- |
Phytosulfokine 3 precursor, putative isoform 2 [Theobroma cacao] |