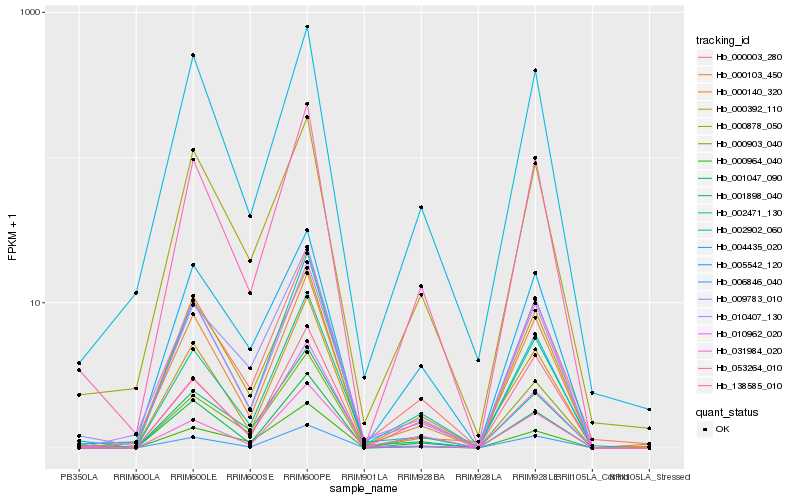
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_053264_010 |
0.0 |
- |
- |
beta glucosidase [Hevea brasiliensis] |
| 2 |
Hb_000903_040 |
0.0614782995 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 3 |
Hb_006846_040 |
0.0624928524 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 4 |
Hb_000103_450 |
0.0661283905 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 5 |
Hb_010962_020 |
0.0794886143 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |
| 6 |
Hb_000003_280 |
0.0808971442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001047_090 |
0.082014417 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |
| 8 |
Hb_002471_130 |
0.0869885343 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |
| 9 |
Hb_031984_020 |
0.089943321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000140_320 |
0.0930838953 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 11 |
Hb_009783_010 |
0.0955576348 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 12 |
Hb_000878_050 |
0.0957663246 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 13 |
Hb_010407_130 |
0.0978692958 |
- |
- |
hypothetical protein POPTR_0008s16680g [Populus trichocarpa] |
| 14 |
Hb_138585_010 |
0.099297012 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 15 |
Hb_005542_120 |
0.1007469923 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040 [Jatropha curcas] |
| 16 |
Hb_001898_040 |
0.1013874126 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 17 |
Hb_004435_020 |
0.1013883207 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 18 |
Hb_000392_110 |
0.1022740468 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 19 |
Hb_002902_060 |
0.1058072744 |
- |
- |
PREDICTED: uncharacterized protein LOC105639824 [Jatropha curcas] |
| 20 |
Hb_000964_040 |
0.1070053383 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER isoform X2 [Jatropha curcas] |