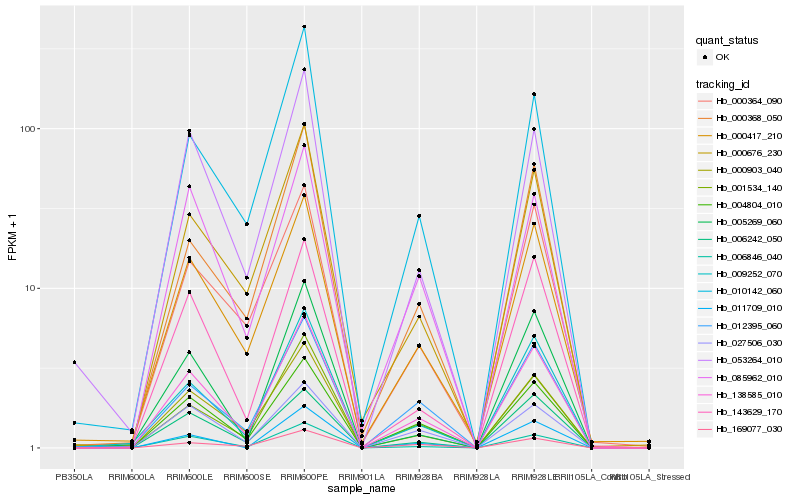
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_138585_010 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 2 |
Hb_004804_010 |
0.0592984406 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 3 |
Hb_006846_040 |
0.0730629482 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 4 |
Hb_001534_140 |
0.0795403319 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105642310 [Jatropha curcas] |
| 5 |
Hb_000417_210 |
0.0835349743 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_009252_070 |
0.0867382093 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 7 |
Hb_000903_040 |
0.0879754033 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 8 |
Hb_012395_060 |
0.0912781157 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 9 |
Hb_000676_230 |
0.0917731355 |
- |
- |
PREDICTED: beta-galactosidase 3 [Jatropha curcas] |
| 10 |
Hb_143629_170 |
0.095129919 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 11 |
Hb_006242_050 |
0.0967248919 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 12 |
Hb_053264_010 |
0.099297012 |
- |
- |
beta glucosidase [Hevea brasiliensis] |
| 13 |
Hb_027506_030 |
0.099650592 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_011709_010 |
0.1015181588 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 15 |
Hb_000368_050 |
0.1024493076 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 16 |
Hb_085962_010 |
0.1036574662 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 17 |
Hb_169077_030 |
0.1062260675 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein RF9 [Jatropha curcas] |
| 18 |
Hb_010142_060 |
0.106440861 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 19 |
Hb_000364_090 |
0.1088736264 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005269_060 |
0.1142414471 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |