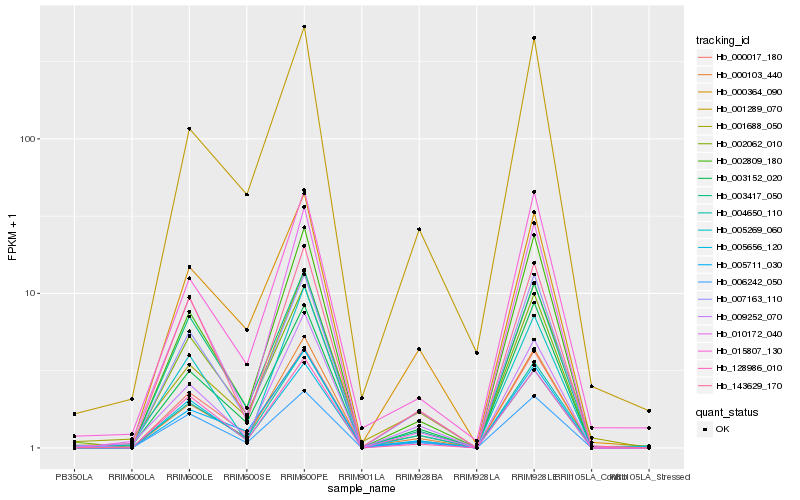
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004650_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s08760g [Populus trichocarpa] |
| 2 |
Hb_002809_180 |
0.0713234199 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 3 |
Hb_007163_110 |
0.0747091502 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 4 |
Hb_003152_020 |
0.0775338728 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 5 |
Hb_000017_180 |
0.0786016983 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 6 |
Hb_001289_070 |
0.0811834696 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 7 |
Hb_128986_010 |
0.0828515635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_005711_030 |
0.083986776 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 9 |
Hb_143629_170 |
0.0841103372 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 10 |
Hb_000103_440 |
0.0869375684 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 11 |
Hb_009252_070 |
0.0877650883 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 12 |
Hb_010172_040 |
0.0881212786 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_015807_130 |
0.0890427593 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003417_050 |
0.0897393949 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_002062_010 |
0.0908375183 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 16 |
Hb_000364_090 |
0.0932488922 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001688_050 |
0.0937227748 |
- |
- |
PREDICTED: reticulon-like protein B9 [Jatropha curcas] |
| 18 |
Hb_005656_120 |
0.0956479079 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 19 |
Hb_005269_060 |
0.0966543759 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 20 |
Hb_006242_050 |
0.0979069472 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |