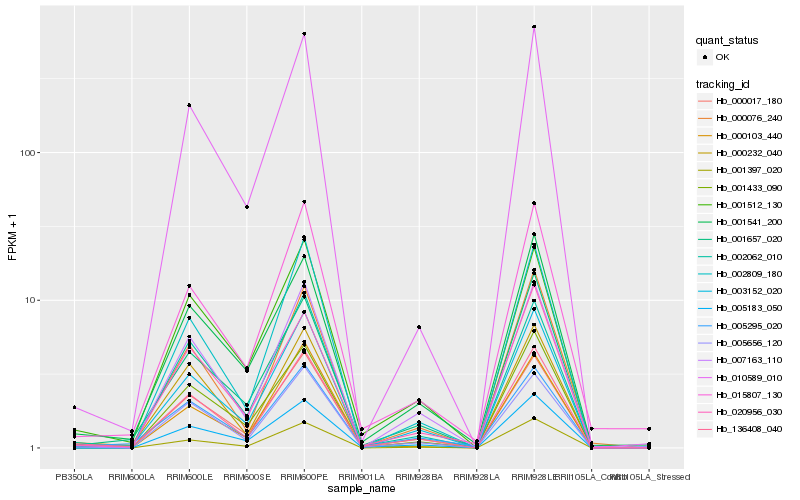
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_136408_040 |
0.0 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 2 |
Hb_003152_020 |
0.0655259964 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 3 |
Hb_007163_110 |
0.0709809548 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 4 |
Hb_002062_010 |
0.0737642939 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 5 |
Hb_001512_130 |
0.0756236014 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 6 |
Hb_000017_180 |
0.0804869065 |
- |
- |
hypothetical protein JCGZ_20551 [Jatropha curcas] |
| 7 |
Hb_000076_240 |
0.0813471096 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 8 |
Hb_005295_020 |
0.082955679 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 9 |
Hb_005656_120 |
0.0836857817 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 10 |
Hb_001433_090 |
0.0843603717 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 11 |
Hb_000103_440 |
0.0861714652 |
- |
- |
hypothetical protein POPTR_0008s15900g [Populus trichocarpa] |
| 12 |
Hb_000232_040 |
0.0866265102 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 isoform X2 [Populus euphratica] |
| 13 |
Hb_005183_050 |
0.0892930379 |
- |
- |
hypothetical protein RCOM_1590900 [Ricinus communis] |
| 14 |
Hb_001541_200 |
0.0894087611 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At1g67900-like isoform X1 [Citrus sinensis] |
| 15 |
Hb_015807_130 |
0.0903619346 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_010589_010 |
0.0917218803 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 17 |
Hb_001657_020 |
0.0929486323 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 18 |
Hb_002809_180 |
0.0980841076 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 19 |
Hb_001397_020 |
0.0981363231 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 20 |
Hb_020956_030 |
0.0998079343 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |