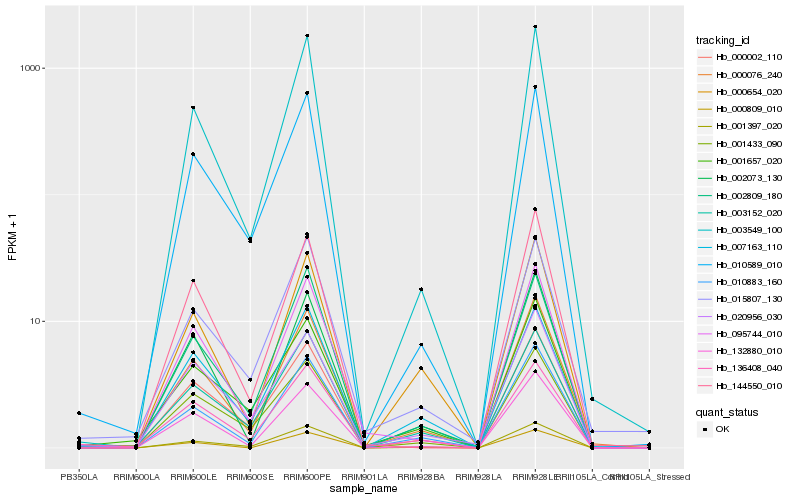
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000076_240 |
0.0 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 2 |
Hb_003549_100 |
0.0707736963 |
- |
- |
PREDICTED: uncharacterized protein LOC105647410 [Jatropha curcas] |
| 3 |
Hb_010883_160 |
0.0722815282 |
- |
- |
PREDICTED: pathogenesis-related protein 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_132880_010 |
0.0746177226 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 5 |
Hb_001397_020 |
0.0755518733 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 6 |
Hb_003152_020 |
0.0813209131 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Jatropha curcas] |
| 7 |
Hb_136408_040 |
0.0813471096 |
- |
- |
PREDICTED: protein NDR1 [Jatropha curcas] |
| 8 |
Hb_002073_130 |
0.0829900834 |
- |
- |
PREDICTED: uncharacterized protein LOC100246830 [Vitis vinifera] |
| 9 |
Hb_095744_010 |
0.0840730351 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_010589_010 |
0.0865535004 |
- |
- |
PREDICTED: classical arabinogalactan protein 9-like [Jatropha curcas] |
| 11 |
Hb_001657_020 |
0.0883814134 |
- |
- |
heat shock family protein [Populus trichocarpa] |
| 12 |
Hb_007163_110 |
0.0890361158 |
- |
- |
PREDICTED: kinesin-like protein KIF19 [Jatropha curcas] |
| 13 |
Hb_000654_020 |
0.0896581613 |
- |
- |
hypothetical protein CICLE_v10006262mg [Citrus clementina] |
| 14 |
Hb_000002_110 |
0.0913066494 |
- |
- |
hypothetical protein EUGRSUZ_B02140 [Eucalyptus grandis] |
| 15 |
Hb_001433_090 |
0.0932657751 |
- |
- |
Pectinesterase PPE8B precursor, putative [Ricinus communis] |
| 16 |
Hb_020956_030 |
0.0946760442 |
- |
- |
PREDICTED: swi5-dependent recombination DNA repair protein 1 homolog [Jatropha curcas] |
| 17 |
Hb_015807_130 |
0.0951644263 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_144550_010 |
0.0979125462 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_002809_180 |
0.0982151964 |
- |
- |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 20 |
Hb_000809_010 |
0.0983008328 |
- |
- |
hypothetical protein JCGZ_12342 [Jatropha curcas] |