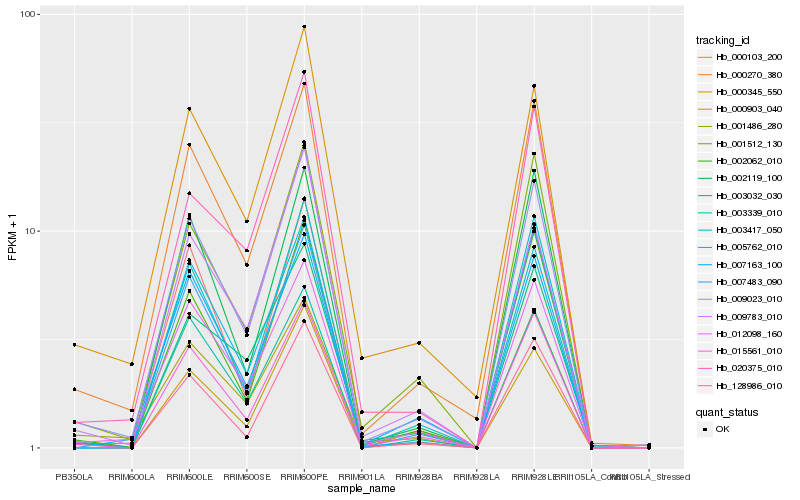
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009023_010 |
0.0 |
- |
- |
glycosyl hydrolase family 31 family protein [Populus trichocarpa] |
| 2 |
Hb_000270_380 |
0.062716682 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 3 |
Hb_015561_010 |
0.0651581872 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005762_010 |
0.0725862234 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_002062_010 |
0.0734044666 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 6 |
Hb_012098_160 |
0.0740922741 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 7 |
Hb_001512_130 |
0.0787793936 |
- |
- |
PREDICTED: uncharacterized protein LOC105633633 [Jatropha curcas] |
| 8 |
Hb_003417_050 |
0.0885756842 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_128986_010 |
0.0889861256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001486_280 |
0.0890366806 |
- |
- |
hypothetical protein RCOM_1321910 [Ricinus communis] |
| 11 |
Hb_000103_200 |
0.089989555 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 12 |
Hb_009783_010 |
0.0917773705 |
- |
- |
PREDICTED: sulfated surface glycoprotein 185-like [Populus euphratica] |
| 13 |
Hb_003032_030 |
0.0918310689 |
transcription factor |
TF Family: G2-like |
PREDICTED: uncharacterized protein LOC105639296 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007163_100 |
0.0925887932 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 15 |
Hb_000345_550 |
0.0936991984 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 16 |
Hb_000903_040 |
0.0952880533 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 17 |
Hb_020375_010 |
0.0957991461 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 18 |
Hb_002119_100 |
0.0967360721 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 19 |
Hb_003339_010 |
0.098170596 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 20 |
Hb_007483_090 |
0.0987541825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |