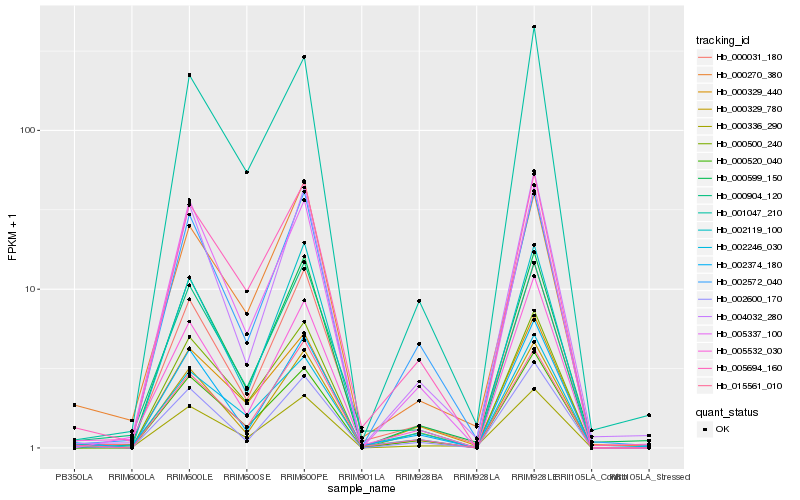
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000336_290 |
0.0 |
- |
- |
leucine-rich repeat protein, putative [Ricinus communis] |
| 2 |
Hb_000500_240 |
0.0838354636 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000270_380 |
0.0856654892 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 4 |
Hb_000031_180 |
0.0878792911 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 5 |
Hb_000599_150 |
0.0936239483 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 6 |
Hb_005694_160 |
0.0948343938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000329_440 |
0.0988444279 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 8 |
Hb_000904_120 |
0.0994662397 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001047_210 |
0.0998045204 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 10 |
Hb_005532_030 |
0.1001295113 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000520_040 |
0.1007525278 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 12 |
Hb_000329_780 |
0.1016167767 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 13 |
Hb_002246_030 |
0.1039457029 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 14 |
Hb_005337_100 |
0.10428682 |
- |
- |
PREDICTED: uncharacterized protein LOC105629349 [Jatropha curcas] |
| 15 |
Hb_002374_180 |
0.1042891453 |
- |
- |
PREDICTED: E3 ubiquitin ligase BIG BROTHER-related [Jatropha curcas] |
| 16 |
Hb_004032_280 |
0.1054326102 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 17 |
Hb_002600_170 |
0.1063058135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002119_100 |
0.1077974455 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 19 |
Hb_002572_040 |
0.1081654664 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 20 |
Hb_015561_010 |
0.1086775149 |
- |
- |
conserved hypothetical protein [Ricinus communis] |