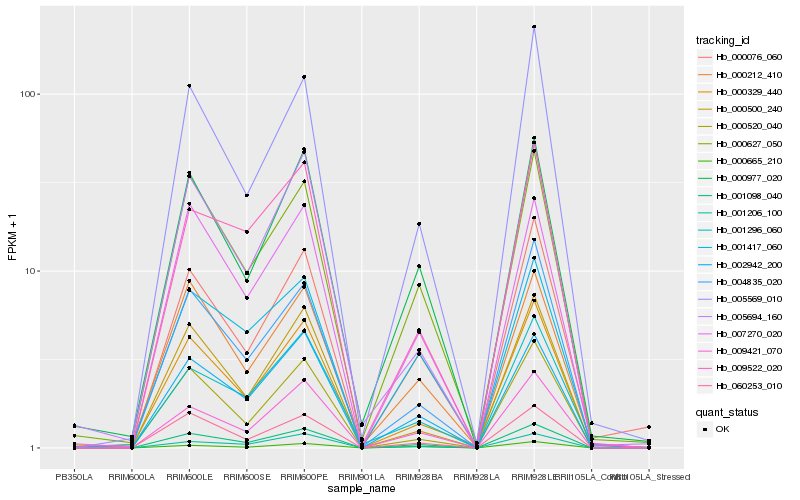
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001098_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 2 |
Hb_002942_200 |
0.0547482945 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH137 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000329_440 |
0.0649102751 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 4 |
Hb_009421_070 |
0.0661383368 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 5 |
Hb_060253_010 |
0.0770403791 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 6 |
Hb_000627_050 |
0.0803025717 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 7 |
Hb_000977_020 |
0.0805546054 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 8 |
Hb_001206_100 |
0.0836291151 |
- |
- |
hypothetical protein PRUPE_ppa021915mg, partial [Prunus persica] |
| 9 |
Hb_007270_020 |
0.0848592097 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 10 |
Hb_004835_020 |
0.0854447742 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 11 |
Hb_000076_060 |
0.0862950771 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 12 |
Hb_001296_060 |
0.0877481103 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 13 |
Hb_005569_010 |
0.0892429954 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 14 |
Hb_000520_040 |
0.0909695701 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Jatropha curcas] |
| 15 |
Hb_000665_210 |
0.0922394309 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 16 |
Hb_009522_020 |
0.0929359206 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 17 |
Hb_000500_240 |
0.0932133062 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005694_160 |
0.0933905803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000212_410 |
0.0938479754 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 20 |
Hb_001417_060 |
0.0941786221 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |