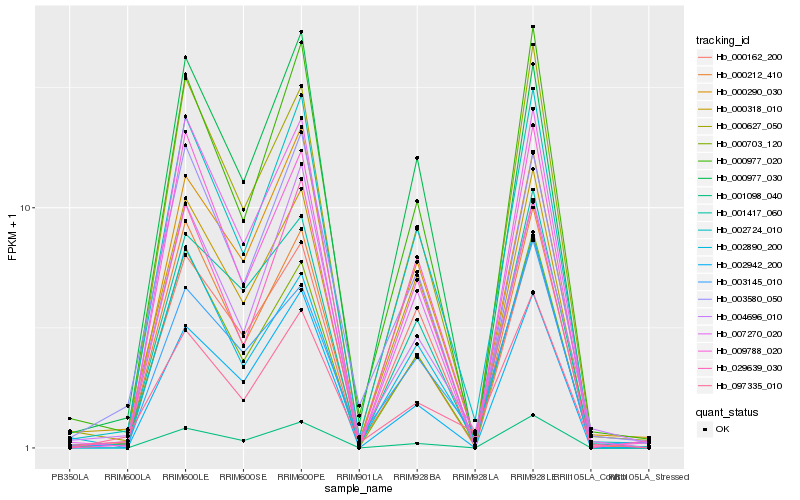
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002724_010 |
0.0 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 2 |
Hb_000977_020 |
0.0607745595 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 3 |
Hb_000703_120 |
0.0676956621 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000212_410 |
0.0700567253 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 5 |
Hb_000318_010 |
0.076859438 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 6 |
Hb_000627_050 |
0.0769597634 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 7 |
Hb_000290_030 |
0.0836415364 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 8 |
Hb_009788_020 |
0.0842005762 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 9 |
Hb_000977_030 |
0.0920234343 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_007270_020 |
0.0934239343 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 11 |
Hb_003580_050 |
0.0957444974 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 12 |
Hb_002890_200 |
0.0970916115 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 13 |
Hb_001417_060 |
0.0972594088 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 14 |
Hb_004696_010 |
0.0974590072 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 15 |
Hb_002942_200 |
0.0978548822 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH137 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000162_200 |
0.099729233 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001098_040 |
0.1002376601 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 18 |
Hb_029639_030 |
0.1062176038 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 19 |
Hb_097335_010 |
0.106635975 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 20 |
Hb_003145_010 |
0.1095963152 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |