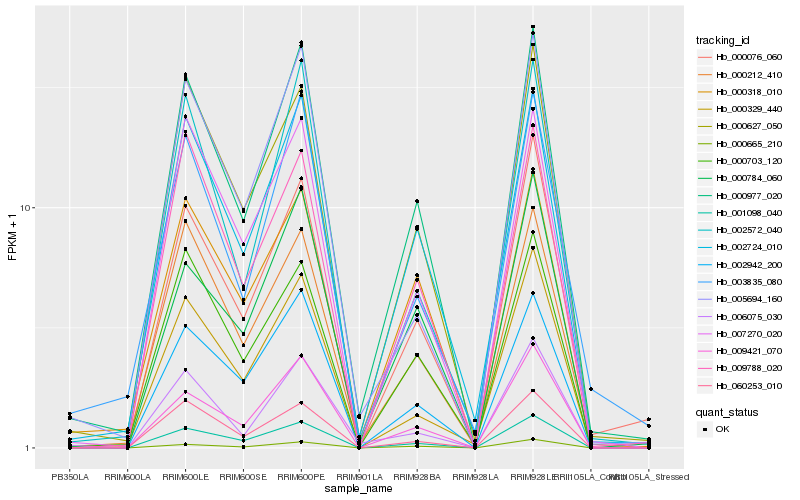
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000977_020 |
0.0 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 2 |
Hb_002724_010 |
0.0607745595 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 3 |
Hb_000627_050 |
0.0749671796 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 4 |
Hb_000212_410 |
0.0765578571 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 5 |
Hb_001098_040 |
0.0805546054 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 6 |
Hb_009421_070 |
0.0896148916 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 7 |
Hb_000703_120 |
0.090847979 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002942_200 |
0.0915598204 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH137 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000076_060 |
0.0928390997 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 10 |
Hb_000318_010 |
0.0928558166 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 11 |
Hb_002572_040 |
0.0936844305 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 12 |
Hb_000665_210 |
0.0956844192 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 13 |
Hb_007270_020 |
0.0967655762 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 14 |
Hb_009788_020 |
0.0991979942 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g06840 [Jatropha curcas] |
| 15 |
Hb_005694_160 |
0.0998730519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000784_060 |
0.1008277744 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003835_080 |
0.1010964052 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_006075_030 |
0.1024460134 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 19 |
Hb_060253_010 |
0.1042891198 |
- |
- |
hypothetical protein JCGZ_14707 [Jatropha curcas] |
| 20 |
Hb_000329_440 |
0.1060222826 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |