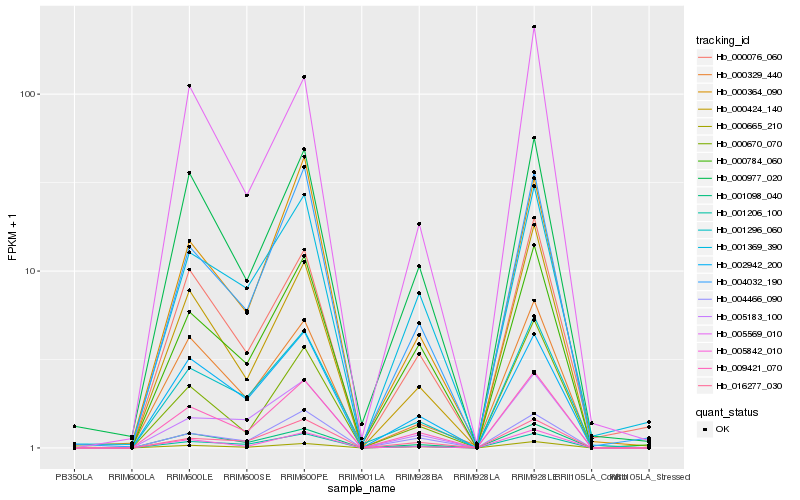
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009421_070 |
0.0 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 2 |
Hb_000665_210 |
0.0598198097 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 3 |
Hb_001206_100 |
0.0660793024 |
- |
- |
hypothetical protein PRUPE_ppa021915mg, partial [Prunus persica] |
| 4 |
Hb_001098_040 |
0.0661383368 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 5 |
Hb_000784_060 |
0.0711542683 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005842_010 |
0.0728259575 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 7 |
Hb_004032_190 |
0.0748692726 |
- |
- |
PREDICTED: probable cellulose synthase A catalytic subunit 3 [UDP-forming] isoform X1 [Jatropha curcas] |
| 8 |
Hb_000424_140 |
0.0803718751 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_002942_200 |
0.0828755745 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH137 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001296_060 |
0.0836148701 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 11 |
Hb_016277_030 |
0.0843294073 |
- |
- |
hypothetical protein JCGZ_16973 [Jatropha curcas] |
| 12 |
Hb_004466_090 |
0.0884536962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000977_020 |
0.0896148916 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 14 |
Hb_000329_440 |
0.0932219984 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 15 |
Hb_000076_060 |
0.0932309521 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 16 |
Hb_000670_070 |
0.0982884692 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 17 |
Hb_005183_100 |
0.0992207675 |
- |
- |
hypothetical protein CICLE_v10002898mg [Citrus clementina] |
| 18 |
Hb_001369_390 |
0.1037126775 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 19 |
Hb_005569_010 |
0.1068835699 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000364_090 |
0.1075929263 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X2 [Jatropha curcas] |