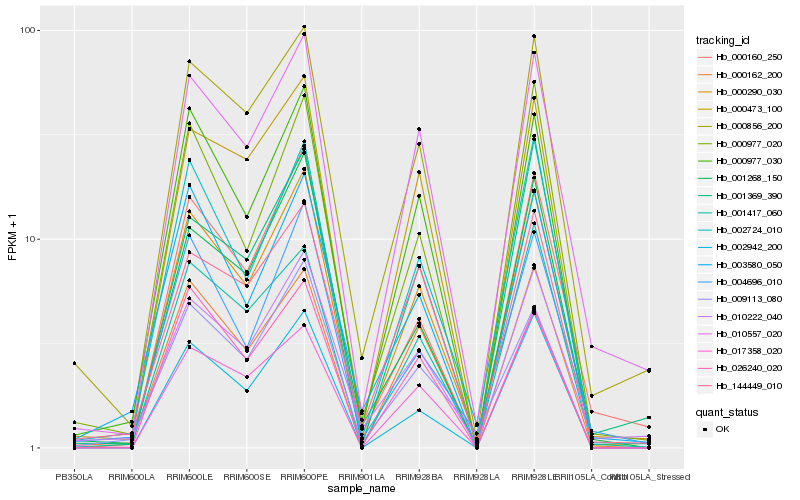
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000290_030 |
0.0 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 2 |
Hb_010557_020 |
0.061407549 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 3 |
Hb_000977_030 |
0.0797773547 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000856_200 |
0.0821574542 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 5 |
Hb_002724_010 |
0.0836415364 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 6 |
Hb_000160_250 |
0.0844504275 |
- |
- |
Uncharacterized protein TCM_019235 [Theobroma cacao] |
| 7 |
Hb_000473_100 |
0.089724589 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 8 |
Hb_010222_040 |
0.0899325704 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |
| 9 |
Hb_002942_200 |
0.0925097582 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH137 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004696_010 |
0.100060447 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 11 |
Hb_000162_200 |
0.1008554346 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001417_060 |
0.1011252346 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 13 |
Hb_001369_390 |
0.1022322537 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 14 |
Hb_001268_150 |
0.1026610578 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_009113_080 |
0.1049844484 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 16 |
Hb_003580_050 |
0.1084768312 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 17 |
Hb_026240_020 |
0.1086800843 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 18 |
Hb_000977_020 |
0.1087054991 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 19 |
Hb_017358_020 |
0.1093550151 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 20 |
Hb_144449_010 |
0.1116204796 |
- |
- |
Potassium transporter, putative [Ricinus communis] |