| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
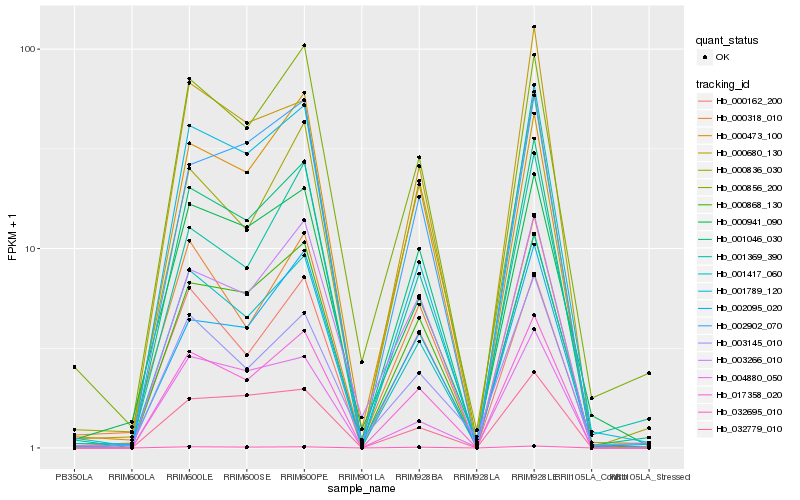
Hb_001046_030 |
0.0 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_017358_020 |
0.0460833568 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 3 |
Hb_001417_060 |
0.0535238966 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 4 |
Hb_000868_130 |
0.0544408413 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 5 |
Hb_003266_010 |
0.0878631664 |
- |
- |
hypothetical protein JCGZ_21945 [Jatropha curcas] |
| 6 |
Hb_032695_010 |
0.0906860698 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Vitis vinifera] |
| 7 |
Hb_000856_200 |
0.0910661115 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 8 |
Hb_000473_100 |
0.0925803889 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_001789_120 |
0.09322593 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_002902_070 |
0.0933903546 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g67385 [Jatropha curcas] |
| 11 |
Hb_002095_020 |
0.0974312115 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_001369_390 |
0.0981009802 |
- |
- |
Glycerol-3-phosphate transporter, putative [Ricinus communis] |
| 13 |
Hb_032779_010 |
0.0984658902 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000680_130 |
0.100028469 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 15 |
Hb_000941_090 |
0.1002676253 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 16 |
Hb_000162_200 |
0.1065692195 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000318_010 |
0.1068409394 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 18 |
Hb_004880_050 |
0.109436719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003145_010 |
0.109864567 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 20 |
Hb_000836_030 |
0.1108116196 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |