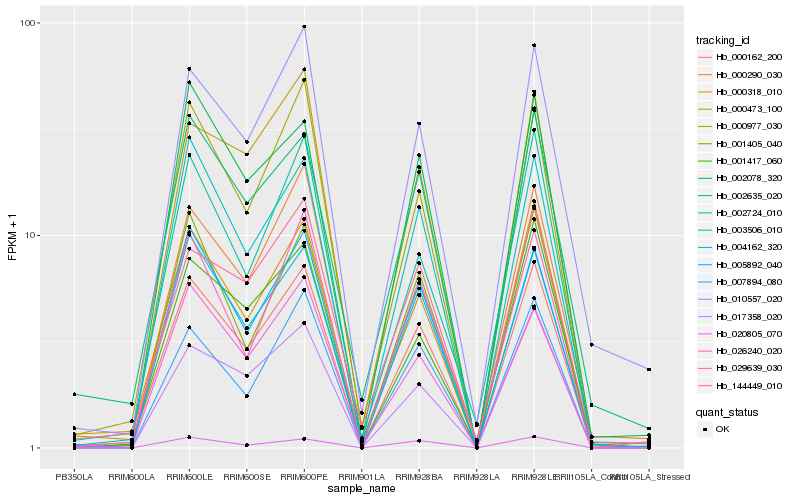
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000162_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000318_010 |
0.0649409615 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 3 |
Hb_003506_010 |
0.0766622249 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 4 |
Hb_004162_320 |
0.0817381958 |
- |
- |
PREDICTED: uncharacterized protein LOC105628009 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000977_030 |
0.0828398259 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_005892_040 |
0.0882467524 |
- |
- |
hypothetical protein JCGZ_02368 [Jatropha curcas] |
| 7 |
Hb_007894_080 |
0.0912027745 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_017358_020 |
0.0935136165 |
- |
- |
hypothetical protein POPTR_0014s16490g [Populus trichocarpa] |
| 9 |
Hb_001417_060 |
0.0937261386 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 10 |
Hb_020805_070 |
0.0939824956 |
- |
- |
PREDICTED: MLO protein homolog 1-like [Jatropha curcas] |
| 11 |
Hb_001405_040 |
0.0948585932 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 12 |
Hb_010557_020 |
0.095104649 |
- |
- |
Mlo protein [Hevea brasiliensis] |
| 13 |
Hb_002635_020 |
0.0960822444 |
- |
- |
hypothetical protein POPTR_0018s09790g [Populus trichocarpa] |
| 14 |
Hb_002724_010 |
0.099729233 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 15 |
Hb_000473_100 |
0.1004740957 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 16 |
Hb_000290_030 |
0.1008554346 |
- |
- |
phosphoinositide 5-phosphatase, putative [Ricinus communis] |
| 17 |
Hb_144449_010 |
0.1009336109 |
- |
- |
Potassium transporter, putative [Ricinus communis] |
| 18 |
Hb_029639_030 |
0.1020550055 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 19 |
Hb_026240_020 |
0.1022775519 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 20 |
Hb_002078_320 |
0.10304415 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |