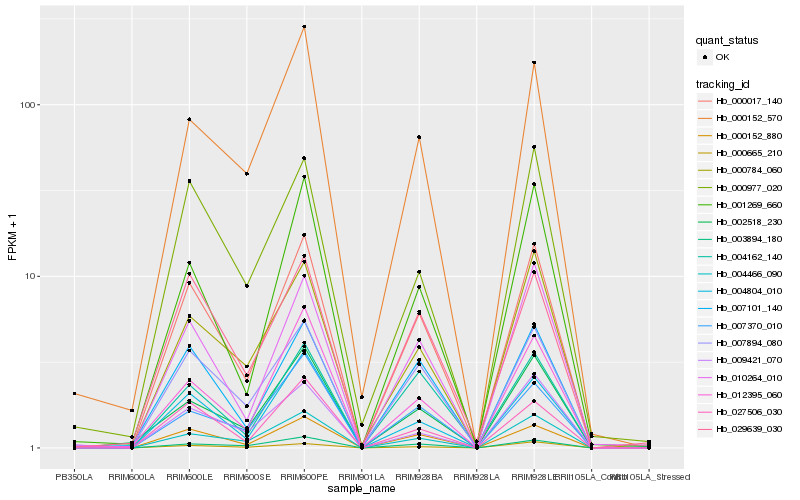
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000017_140 |
0.0 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 2 |
Hb_000152_880 |
0.0664062536 |
- |
- |
- |
| 3 |
Hb_007894_080 |
0.0781242449 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007101_140 |
0.0841101205 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 5 |
Hb_010264_010 |
0.0873441106 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 6 |
Hb_004466_090 |
0.0877016799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_004804_010 |
0.0962094173 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 8 |
Hb_000784_060 |
0.0979092418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003894_180 |
0.1024557136 |
- |
- |
PREDICTED: uncharacterized protein LOC104245193, partial [Nicotiana sylvestris] |
| 10 |
Hb_002518_230 |
0.1034125651 |
- |
- |
- |
| 11 |
Hb_000665_210 |
0.1038674613 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 12 |
Hb_027506_030 |
0.1073527549 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 13 |
Hb_029639_030 |
0.1109029162 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 14 |
Hb_001269_660 |
0.1109524002 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 15 |
Hb_000152_570 |
0.1131358435 |
- |
- |
21 kDa protein precursor, putative [Ricinus communis] |
| 16 |
Hb_012395_060 |
0.113535632 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 17 |
Hb_000977_020 |
0.1176100432 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 18 |
Hb_007370_010 |
0.1183516741 |
- |
- |
PREDICTED: indole-3-pyruvate monooxygenase YUCCA6 [Jatropha curcas] |
| 19 |
Hb_009421_070 |
0.118808861 |
- |
- |
PREDICTED: cell number regulator 1-like [Jatropha curcas] |
| 20 |
Hb_004162_140 |
0.1193826768 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |