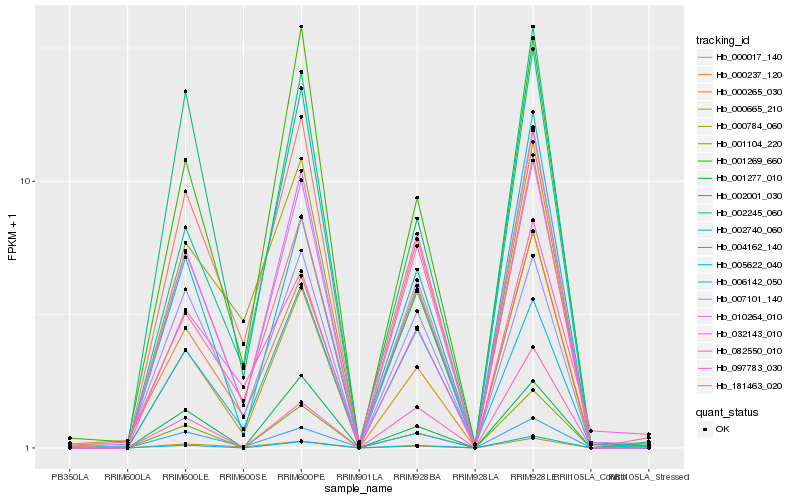
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032143_010 |
0.0 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 2 |
Hb_010264_010 |
0.0764032031 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 3 |
Hb_000265_030 |
0.0874088079 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 4 |
Hb_001104_220 |
0.1000897855 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 5 |
Hb_005622_040 |
0.1061841315 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Jatropha curcas] |
| 6 |
Hb_006142_050 |
0.1097963959 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 7 |
Hb_181463_020 |
0.1193513 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 8 |
Hb_000237_120 |
0.1199882354 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 9 |
Hb_000665_210 |
0.1212446856 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 10 |
Hb_001269_660 |
0.1242485602 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 2 [Jatropha curcas] |
| 11 |
Hb_000017_140 |
0.1282743815 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 12 |
Hb_002740_060 |
0.1295130998 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 13 |
Hb_002245_060 |
0.131065324 |
- |
- |
PREDICTED: kinesin-3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007101_140 |
0.1375374186 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 15 |
Hb_097783_030 |
0.1390229513 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 16 |
Hb_004162_140 |
0.139680289 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 17 |
Hb_082550_010 |
0.1398173378 |
- |
- |
hypothetical protein CISIN_1g0081081mg, partial [Citrus sinensis] |
| 18 |
Hb_000784_060 |
0.1405573315 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002001_030 |
0.1415043274 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 20 |
Hb_001277_010 |
0.1431742931 |
- |
- |
unknown [Populus trichocarpa] |