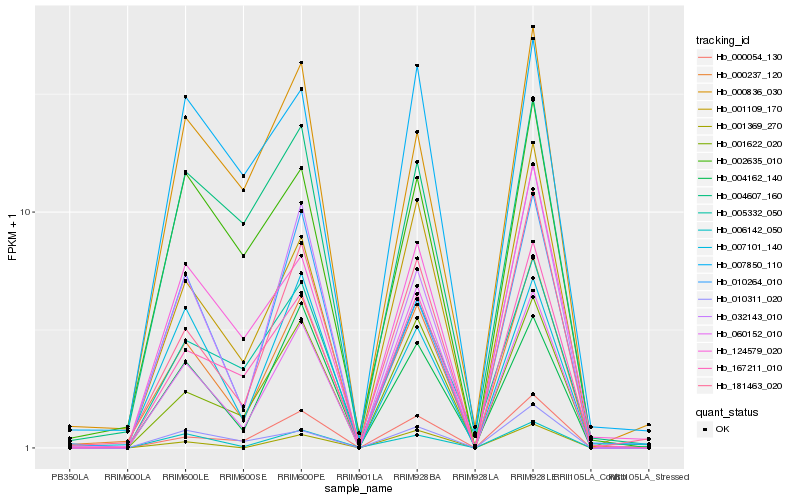
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000237_120 |
0.0 |
- |
- |
PREDICTED: beta-galactosidase 3-like [Jatropha curcas] |
| 2 |
Hb_181463_020 |
0.0843209333 |
- |
- |
hypothetical protein JCGZ_26765 [Jatropha curcas] |
| 3 |
Hb_006142_050 |
0.105785807 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 4 |
Hb_167211_010 |
0.1152979213 |
- |
- |
PREDICTED: glutamate receptor 3.4-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001109_170 |
0.1183189452 |
- |
- |
PREDICTED: urea-proton symporter DUR3 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001622_020 |
0.1183779817 |
- |
- |
- |
| 7 |
Hb_032143_010 |
0.1199882354 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 8 |
Hb_000054_130 |
0.1207596514 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 9 |
Hb_002635_010 |
0.1349712429 |
- |
- |
homogentisate phytyl transferase [Hevea brasiliensis] |
| 10 |
Hb_060152_010 |
0.1350297107 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 11 |
Hb_004162_140 |
0.1357717034 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 12 |
Hb_124579_020 |
0.1364362426 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 13 |
Hb_007101_140 |
0.1384114706 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 14 |
Hb_010311_020 |
0.1395504659 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 15 |
Hb_000836_030 |
0.1459484852 |
- |
- |
ubiquitin-conjugating enzyme h, putative [Ricinus communis] |
| 16 |
Hb_005332_050 |
0.1467385725 |
- |
- |
hypothetical protein POPTR_0017s10650g [Populus trichocarpa] |
| 17 |
Hb_010264_010 |
0.1481963084 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 18 |
Hb_001369_270 |
0.1488246753 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_004607_160 |
0.1498372632 |
- |
- |
PREDICTED: uncharacterized protein LOC105134931 [Populus euphratica] |
| 20 |
Hb_007850_110 |
0.1524026501 |
- |
- |
MADS-box transcription factor 3 [Hevea brasiliensis] |