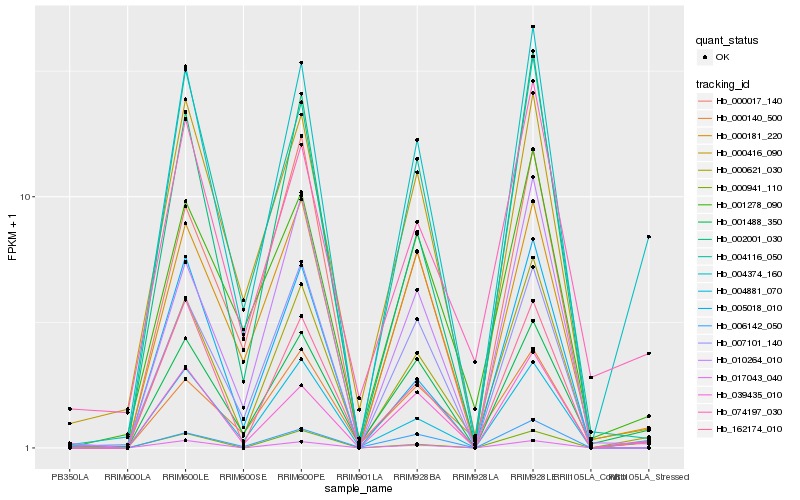
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000621_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002001_030 |
0.0975222682 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 3 |
Hb_004881_070 |
0.1057779554 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_162174_010 |
0.109038397 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_010264_010 |
0.115009585 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 6 |
Hb_004374_160 |
0.1173043768 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 7 |
Hb_004116_050 |
0.1188420241 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 8 |
Hb_039435_010 |
0.1245636672 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 9 |
Hb_006142_050 |
0.1268984871 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 10 |
Hb_000140_500 |
0.1300485995 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_005018_010 |
0.1328474324 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 12 |
Hb_000416_090 |
0.1331617433 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 13 |
Hb_007101_140 |
0.1339279517 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 14 |
Hb_001488_350 |
0.1346277306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001278_090 |
0.134982679 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 16 |
Hb_017043_040 |
0.138680476 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 17 |
Hb_000181_220 |
0.1389297419 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000017_140 |
0.1438318063 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 19 |
Hb_000941_110 |
0.1452097566 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 20 |
Hb_074197_030 |
0.1472250478 |
- |
- |
unnamed protein product [Vitis vinifera] |