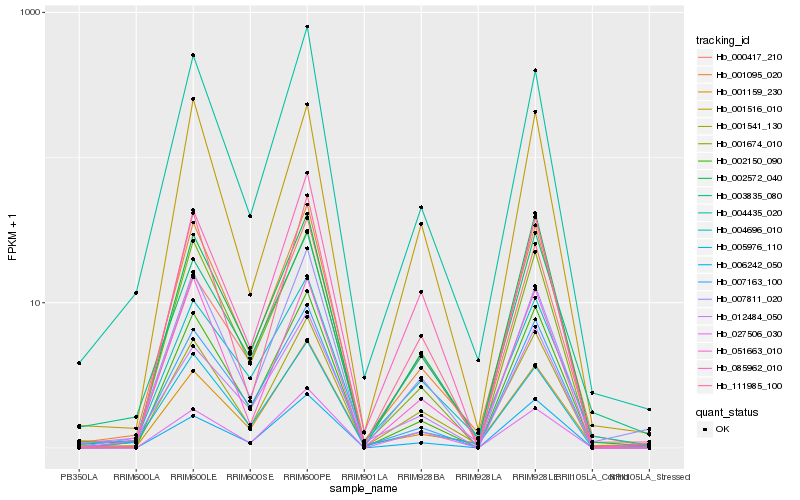
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001541_130 |
0.0 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 2 |
Hb_001095_020 |
0.073989413 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 3 |
Hb_002572_040 |
0.0749262642 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 4 |
Hb_111985_100 |
0.0816371707 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 5 |
Hb_002150_090 |
0.0850394536 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 6 |
Hb_007811_020 |
0.0913196322 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 7 |
Hb_004435_020 |
0.0941073672 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 8 |
Hb_005976_110 |
0.0950475892 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 9 |
Hb_004696_010 |
0.0954835602 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 10 |
Hb_000417_210 |
0.0956698096 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_051663_010 |
0.0963283003 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_001674_010 |
0.0989195673 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 13 |
Hb_007163_100 |
0.0990467449 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 14 |
Hb_001516_010 |
0.0998186869 |
- |
- |
PREDICTED: guanylate kinase 2 [Jatropha curcas] |
| 15 |
Hb_001159_230 |
0.1020646884 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 16 |
Hb_085962_010 |
0.1024343158 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 17 |
Hb_006242_050 |
0.1028913907 |
- |
- |
Osmotin precursor, putative [Ricinus communis] |
| 18 |
Hb_012484_050 |
0.1036909883 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 19 |
Hb_027506_030 |
0.1056223357 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 20 |
Hb_003835_080 |
0.1064201755 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |