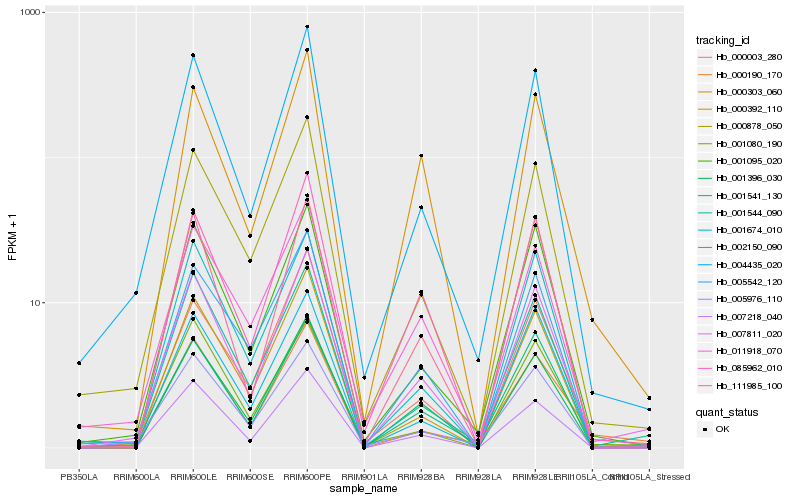
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007811_020 |
0.0 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 2 |
Hb_004435_020 |
0.0719963944 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 3 |
Hb_001544_090 |
0.0798891049 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 4 |
Hb_007218_040 |
0.0811083555 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 5 |
Hb_000878_050 |
0.0843398111 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 6 |
Hb_000392_110 |
0.0886901461 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001541_130 |
0.0913196322 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 8 |
Hb_005976_110 |
0.0914624215 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 9 |
Hb_001095_020 |
0.0921215268 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 10 |
Hb_001396_030 |
0.0940209293 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 11 |
Hb_002150_090 |
0.0980414736 |
- |
- |
PREDICTED: probable inactive receptor kinase At4g23740 [Jatropha curcas] |
| 12 |
Hb_111985_100 |
0.0982147807 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 13 |
Hb_085962_010 |
0.09921469 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 14 |
Hb_005542_120 |
0.0998192826 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040 [Jatropha curcas] |
| 15 |
Hb_000003_280 |
0.0998825168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001674_010 |
0.102196176 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 17 |
Hb_011918_070 |
0.1028206781 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 18 |
Hb_000303_060 |
0.1032585719 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000190_170 |
0.1037110242 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 20 |
Hb_001080_190 |
0.1037815908 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |