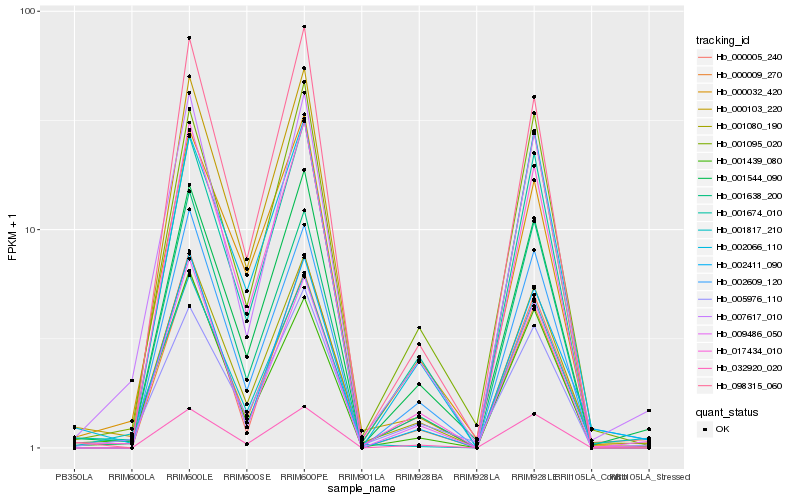
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_190 |
0.0 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007617_010 |
0.0620756696 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH63 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001544_090 |
0.064931424 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 4 |
Hb_001674_010 |
0.0727222848 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 5 |
Hb_001638_200 |
0.0778534197 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 43 [Jatropha curcas] |
| 6 |
Hb_017434_010 |
0.0782694232 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 7 |
Hb_000009_270 |
0.0813441602 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 8 |
Hb_001095_020 |
0.0836578069 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 9 |
Hb_002609_120 |
0.0853921349 |
- |
- |
hypothetical protein POPTR_0001s25660g [Populus trichocarpa] |
| 10 |
Hb_000005_240 |
0.0875770416 |
- |
- |
Shugoshin-1, putative [Ricinus communis] |
| 11 |
Hb_032920_020 |
0.0891509172 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 12 |
Hb_005976_110 |
0.0907693791 |
- |
- |
PREDICTED: uncharacterized protein LOC105639708 [Jatropha curcas] |
| 13 |
Hb_001439_080 |
0.090981664 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 14 |
Hb_009486_050 |
0.0929663324 |
- |
- |
PREDICTED: cyclin-U4-1 [Jatropha curcas] |
| 15 |
Hb_002411_090 |
0.0945834707 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL69 [Jatropha curcas] |
| 16 |
Hb_000103_220 |
0.0949409802 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |
| 17 |
Hb_001817_210 |
0.0949811165 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002066_110 |
0.0950747847 |
- |
- |
PREDICTED: calmodulin-like protein 1 [Jatropha curcas] |
| 19 |
Hb_098315_060 |
0.0954572251 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 20 |
Hb_000032_420 |
0.0968260233 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |