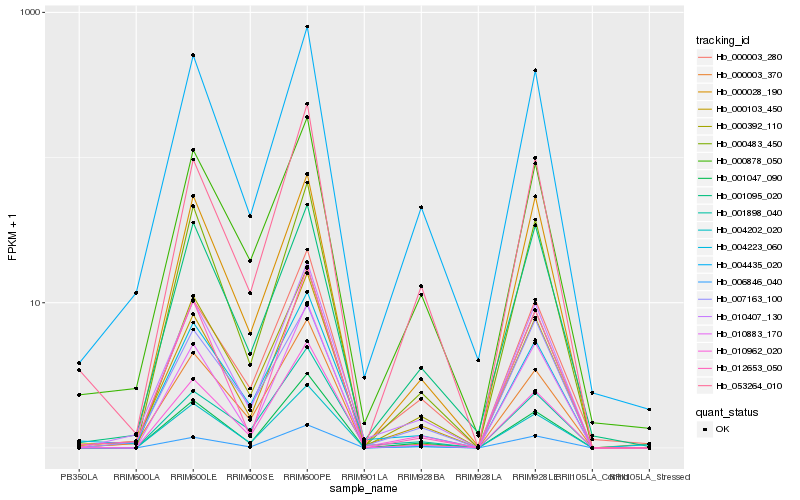
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010407_130 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s16680g [Populus trichocarpa] |
| 2 |
Hb_000103_450 |
0.0461894618 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 3 |
Hb_004435_020 |
0.0712893798 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 4 |
Hb_006846_040 |
0.0842304695 |
- |
- |
PREDICTED: uncharacterized protein LOC105648687 [Jatropha curcas] |
| 5 |
Hb_000003_280 |
0.08689523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000392_110 |
0.087277741 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_000483_450 |
0.0887607887 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 8 |
Hb_000878_050 |
0.0899712864 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 9 |
Hb_004202_020 |
0.0903797067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_010962_020 |
0.0908490512 |
transcription factor |
TF Family: OFP |
hypothetical protein RCOM_0815310 [Ricinus communis] |
| 11 |
Hb_001047_090 |
0.092985999 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |
| 12 |
Hb_004223_060 |
0.0940938496 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 13 |
Hb_012653_050 |
0.0943143786 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 14 |
Hb_007163_100 |
0.0946291919 |
- |
- |
BnaC03g52280D [Brassica napus] |
| 15 |
Hb_000028_190 |
0.0949053977 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 16 |
Hb_053264_010 |
0.0978692958 |
- |
- |
beta glucosidase [Hevea brasiliensis] |
| 17 |
Hb_000003_370 |
0.0981386971 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 18 |
Hb_010883_170 |
0.1007936151 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 19 |
Hb_001095_020 |
0.1021511326 |
- |
- |
PREDICTED: F-box protein At2g26850-like [Jatropha curcas] |
| 20 |
Hb_001898_040 |
0.1035199954 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |