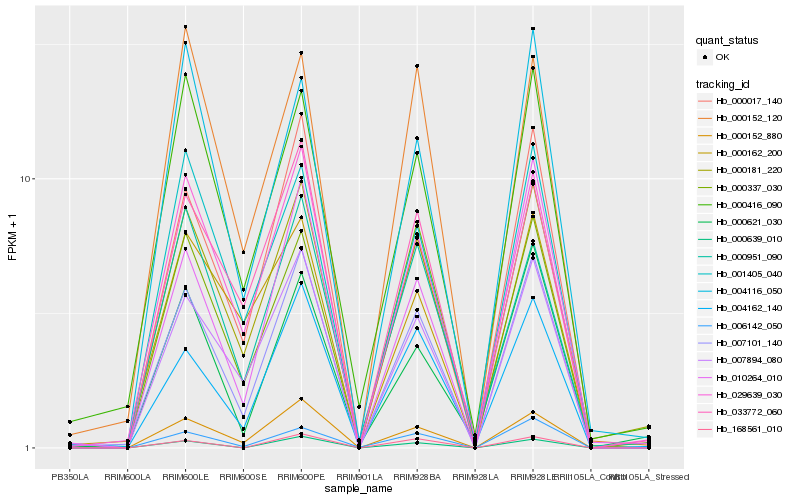
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007101_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 2 |
Hb_000152_880 |
0.074994901 |
- |
- |
- |
| 3 |
Hb_007894_080 |
0.0826691998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000017_140 |
0.0841101205 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 5 |
Hb_029639_030 |
0.0927790996 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |
| 6 |
Hb_000181_220 |
0.0972997881 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_004162_140 |
0.0987629214 |
- |
- |
PREDICTED: serine carboxypeptidase-like 51 [Jatropha curcas] |
| 8 |
Hb_010264_010 |
0.108073692 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 9 |
Hb_006142_050 |
0.1110286522 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 10 |
Hb_001405_040 |
0.1146198543 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 11 |
Hb_033772_060 |
0.1163029532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000639_010 |
0.1190226351 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_09727 [Jatropha curcas] |
| 13 |
Hb_000416_090 |
0.1212882825 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 14 |
Hb_004116_050 |
0.1320688797 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 15 |
Hb_000337_030 |
0.1321728143 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_168561_010 |
0.1322205448 |
- |
- |
PREDICTED: notchless protein homolog [Jatropha curcas] |
| 17 |
Hb_000951_090 |
0.1324266787 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 18 |
Hb_000621_030 |
0.1339279517 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000152_120 |
0.1343232153 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 20 |
Hb_000162_200 |
0.1354879519 |
- |
- |
PREDICTED: uncharacterized protein LOC105641759 isoform X2 [Jatropha curcas] |