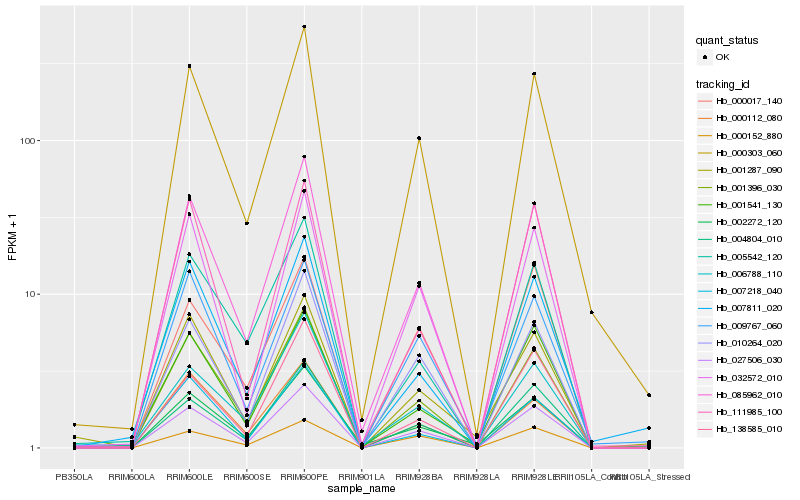
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027506_030 |
0.0 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 2 |
Hb_085962_010 |
0.0456150474 |
- |
- |
hypothetical protein POPTR_0017s11380g [Populus trichocarpa] |
| 3 |
Hb_004804_010 |
0.0537753146 |
- |
- |
PREDICTED: trans-resveratrol di-O-methyltransferase-like [Populus euphratica] |
| 4 |
Hb_001396_030 |
0.0581540165 |
- |
- |
hypothetical protein POPTR_0001s13520g [Populus trichocarpa] |
| 5 |
Hb_010264_020 |
0.062791186 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000303_060 |
0.0648825611 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_032572_010 |
0.0813345198 |
- |
- |
PREDICTED: phospholipase A2-alpha [Jatropha curcas] |
| 8 |
Hb_000152_880 |
0.0956479411 |
- |
- |
- |
| 9 |
Hb_138585_010 |
0.099650592 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor ONAC010 [Jatropha curcas] |
| 10 |
Hb_002272_120 |
0.1028438889 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 11 |
Hb_009767_060 |
0.1041254376 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK9 [Jatropha curcas] |
| 12 |
Hb_001541_130 |
0.1056223357 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 13 |
Hb_000017_140 |
0.1073527549 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 14 |
Hb_000112_080 |
0.1113801705 |
- |
- |
PREDICTED: glutamate receptor 2.1-like [Populus euphratica] |
| 15 |
Hb_005542_120 |
0.1128934265 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At3g28040 [Jatropha curcas] |
| 16 |
Hb_111985_100 |
0.1163671939 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 17 |
Hb_001287_090 |
0.1167153241 |
- |
- |
Endoglucanase 4 [Aegilops tauschii] |
| 18 |
Hb_007811_020 |
0.1175625546 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 19 |
Hb_006788_110 |
0.1181838543 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 20 |
Hb_007218_040 |
0.1187264535 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |