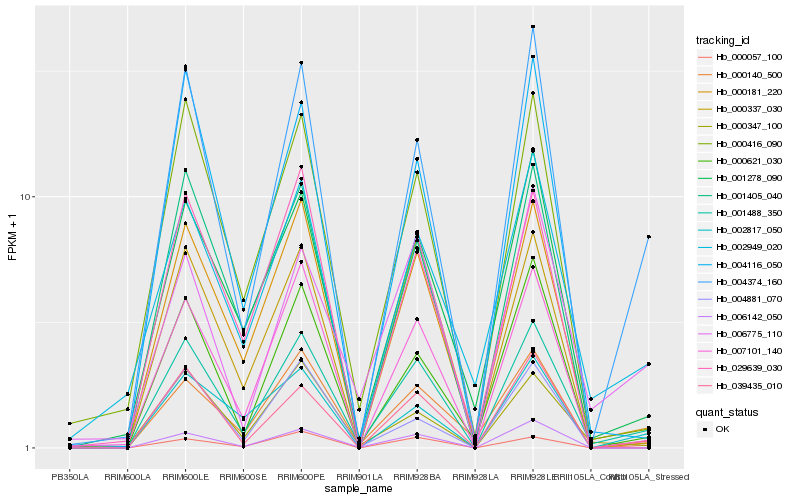
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001488_350 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000181_220 |
0.1008261195 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_004374_160 |
0.1093293735 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 4 |
Hb_039435_010 |
0.1106801677 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 5 |
Hb_000416_090 |
0.1333625437 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 6 |
Hb_000621_030 |
0.1346277306 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001278_090 |
0.1347993658 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 8 |
Hb_000140_500 |
0.1364152163 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_004116_050 |
0.1383273978 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 10 |
Hb_000347_100 |
0.1392114584 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |
| 11 |
Hb_006142_050 |
0.139809645 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 12 |
Hb_004881_070 |
0.1418185994 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000057_100 |
0.1471591546 |
- |
- |
Peroxidase 57 precursor, putative [Ricinus communis] |
| 14 |
Hb_007101_140 |
0.1474639836 |
- |
- |
hypothetical protein JCGZ_21777 [Jatropha curcas] |
| 15 |
Hb_000337_030 |
0.148286988 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_006775_110 |
0.1496329553 |
- |
- |
PREDICTED: uncharacterized protein LOC105649734 [Jatropha curcas] |
| 17 |
Hb_002817_050 |
0.1498245868 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 18 |
Hb_002949_020 |
0.1518148682 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 19 |
Hb_001405_040 |
0.1523082844 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Jatropha curcas] |
| 20 |
Hb_029639_030 |
0.1523794434 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 2-like [Jatropha curcas] |