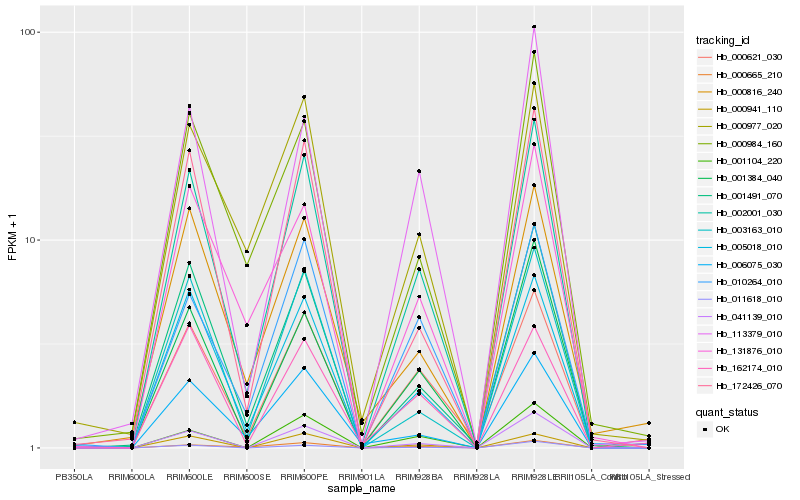
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002001_030 |
0.0 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 2 |
Hb_005018_010 |
0.0772287691 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 3 |
Hb_001491_070 |
0.0793572356 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 17 isoform X1 [Jatropha curcas] |
| 4 |
Hb_041139_010 |
0.0941423962 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 5 |
Hb_172426_070 |
0.0964254049 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000621_030 |
0.0975222682 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001384_040 |
0.1043406681 |
- |
- |
PREDICTED: beta-glucosidase 17-like [Jatropha curcas] |
| 8 |
Hb_003163_010 |
0.1070897657 |
- |
- |
hypothetical protein POPTR_0012s02920g [Populus trichocarpa] |
| 9 |
Hb_000941_110 |
0.1117587069 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 10 |
Hb_010264_010 |
0.1150643695 |
- |
- |
PREDICTED: protein ZINC INDUCED FACILITATOR-LIKE 1-like [Jatropha curcas] |
| 11 |
Hb_113379_010 |
0.1181797145 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 12 |
Hb_001104_220 |
0.1191371449 |
- |
- |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 13 |
Hb_000665_210 |
0.1197523526 |
- |
- |
PREDICTED: synaptotagmin-4 [Jatropha curcas] |
| 14 |
Hb_131876_010 |
0.1205359879 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000984_160 |
0.1216465519 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_011618_010 |
0.1233366244 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g59700 [Jatropha curcas] |
| 17 |
Hb_006075_030 |
0.1235579116 |
- |
- |
PREDICTED: uncharacterized protein LOC105641890 isoform X1 [Jatropha curcas] |
| 18 |
Hb_162174_010 |
0.1244245759 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 19 |
Hb_000816_240 |
0.1247104404 |
- |
- |
PREDICTED: phosphoglucan phosphatase LSF2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000977_020 |
0.1255543197 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |