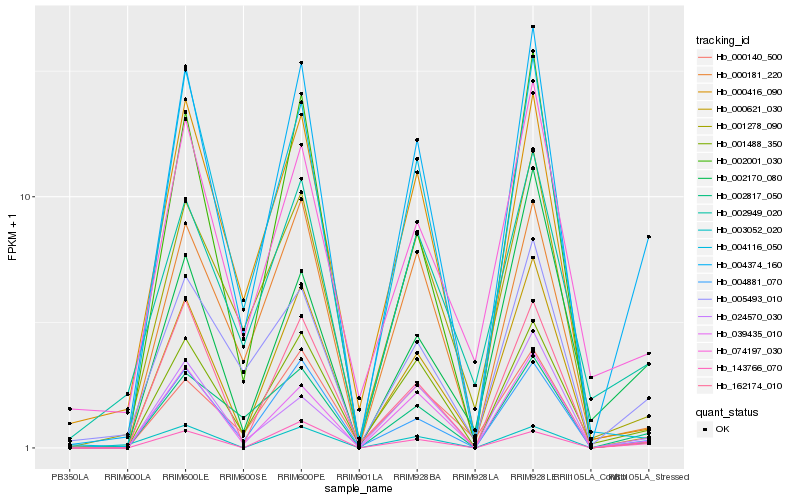
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004374_160 |
0.0 |
transcription factor |
TF Family: bHLH |
hypothetical protein JCGZ_20587 [Jatropha curcas] |
| 2 |
Hb_001488_350 |
0.1093293735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_039435_010 |
0.1102959806 |
- |
- |
hypothetical protein SORBIDRAFT_10g025530 [Sorghum bicolor] |
| 4 |
Hb_000621_030 |
0.1173043768 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004881_070 |
0.1203644391 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_002817_050 |
0.1363248088 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0004s16810g, partial [Populus trichocarpa] |
| 7 |
Hb_005493_010 |
0.1407001186 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_162174_010 |
0.1510693658 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_000181_220 |
0.1533106603 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_024570_030 |
0.1549027903 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_004116_050 |
0.156049997 |
- |
- |
PREDICTED: cytochrome P450 90A1 [Jatropha curcas] |
| 12 |
Hb_001278_090 |
0.1580506351 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_002949_020 |
0.16233929 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0002s23650g [Populus trichocarpa] |
| 14 |
Hb_074197_030 |
0.1639922045 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_143766_070 |
0.1663374369 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 16 |
Hb_000416_090 |
0.166475172 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 17 |
Hb_002001_030 |
0.167088527 |
- |
- |
Plasma membrane ATPase 4 [Gossypium arboreum] |
| 18 |
Hb_002170_080 |
0.1683004499 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 19 |
Hb_000140_500 |
0.170008675 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_003052_020 |
0.172062258 |
- |
- |
PREDICTED: chaperonin 60 subunit beta 4, chloroplastic isoform X1 [Jatropha curcas] |