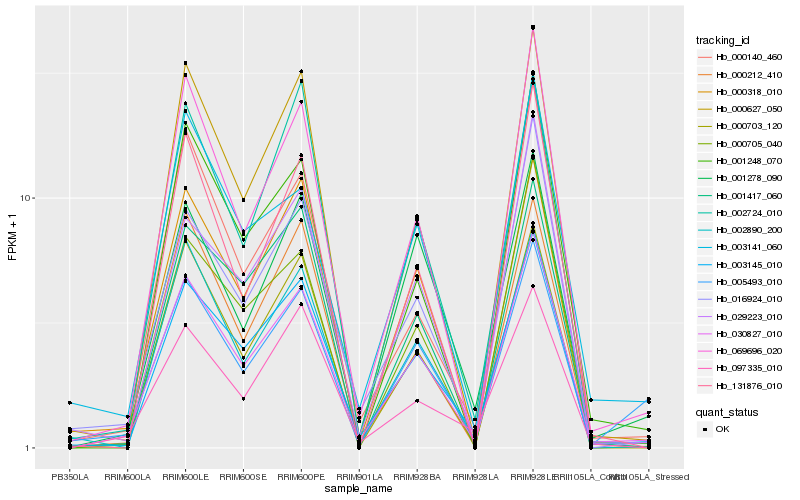
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030827_010 |
0.0 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper ROC6 [Gossypium arboreum] |
| 2 |
Hb_003145_010 |
0.0704188471 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 3 |
Hb_069696_020 |
0.07740674 |
- |
- |
PREDICTED: protochlorophyllide-dependent translocon component 52, chloroplastic-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_000627_050 |
0.1020061109 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 5 |
Hb_016924_010 |
0.1038711316 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000703_120 |
0.1070731014 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001278_090 |
0.109087024 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 8 |
Hb_000140_460 |
0.1095765989 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 9 |
Hb_000705_040 |
0.1108951118 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003141_060 |
0.1117951655 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000318_010 |
0.1118266823 |
- |
- |
calcium-dependent protein kinase 6 [Hevea brasiliensis] |
| 12 |
Hb_001248_070 |
0.1124267135 |
- |
- |
PREDICTED: probable polyamine transporter At3g19553 [Jatropha curcas] |
| 13 |
Hb_001417_060 |
0.1155460011 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 14 |
Hb_029223_010 |
0.1158592828 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 15 |
Hb_002724_010 |
0.1158670059 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 16 |
Hb_002890_200 |
0.1167385855 |
- |
- |
PREDICTED: methionine S-methyltransferase [Jatropha curcas] |
| 17 |
Hb_131876_010 |
0.1169424002 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_097335_010 |
0.1171790192 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 19 |
Hb_000212_410 |
0.1175495963 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 20 |
Hb_005493_010 |
0.1176724917 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |