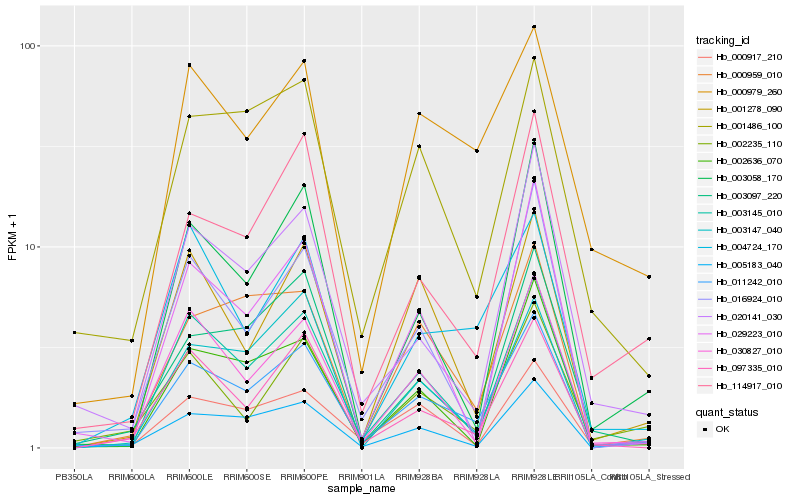
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011242_010 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 2 |
Hb_003145_010 |
0.0966254428 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 3 |
Hb_114917_010 |
0.1314914553 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005183_040 |
0.135232829 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 5 |
Hb_097335_010 |
0.1355250293 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 6 |
Hb_000979_260 |
0.1378559148 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 7 |
Hb_030827_010 |
0.1397332124 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper ROC6 [Gossypium arboreum] |
| 8 |
Hb_000959_010 |
0.1402201025 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000917_210 |
0.1404376043 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003058_170 |
0.1409455662 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001486_100 |
0.1422619433 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 12 |
Hb_020141_030 |
0.1423594027 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 13 |
Hb_001278_090 |
0.1445805658 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 14 |
Hb_002636_070 |
0.1448293678 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 15 |
Hb_004724_170 |
0.1455074834 |
- |
- |
PREDICTED: rho GTPase-activating protein 1 [Jatropha curcas] |
| 16 |
Hb_003147_040 |
0.1460717423 |
- |
- |
PREDICTED: probable calcium-binding protein CML18 [Jatropha curcas] |
| 17 |
Hb_016924_010 |
0.1472575294 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_029223_010 |
0.1479301015 |
- |
- |
PREDICTED: ACT domain-containing protein ACR10 [Jatropha curcas] |
| 19 |
Hb_003097_220 |
0.1520285226 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 20 |
Hb_002235_110 |
0.154366069 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |