| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
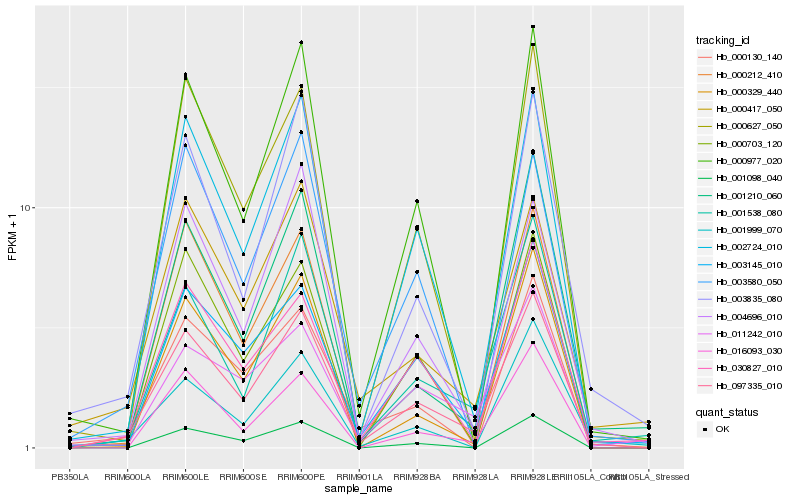
Hb_097335_010 |
0.0 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 2 |
Hb_002724_010 |
0.106635975 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 4-like [Citrus sinensis] |
| 3 |
Hb_001538_080 |
0.1120270326 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_030827_010 |
0.1171790192 |
transcription factor |
TF Family: HB |
Homeobox-leucine zipper ROC6 [Gossypium arboreum] |
| 5 |
Hb_003145_010 |
0.1173114658 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 6 |
Hb_000977_020 |
0.1229048671 |
- |
- |
beta-hexosaminidase, putative [Ricinus communis] |
| 7 |
Hb_003835_080 |
0.1262674527 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000212_410 |
0.1267525876 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 9 |
Hb_016093_030 |
0.130224068 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 10 |
Hb_004696_010 |
0.1314812955 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 11 |
Hb_000627_050 |
0.131802849 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 12 |
Hb_001210_060 |
0.1321142103 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 13 |
Hb_000417_050 |
0.1322270019 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 14 |
Hb_000703_120 |
0.1330412144 |
- |
- |
PREDICTED: uncharacterized protein LOC105635706 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000329_440 |
0.1355245749 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 16 |
Hb_011242_010 |
0.1355250293 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 17 |
Hb_000130_140 |
0.1377690218 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001999_070 |
0.1400249444 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 19 |
Hb_001098_040 |
0.1411106201 |
- |
- |
hypothetical protein JCGZ_00749 [Jatropha curcas] |
| 20 |
Hb_003580_050 |
0.1412645827 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |