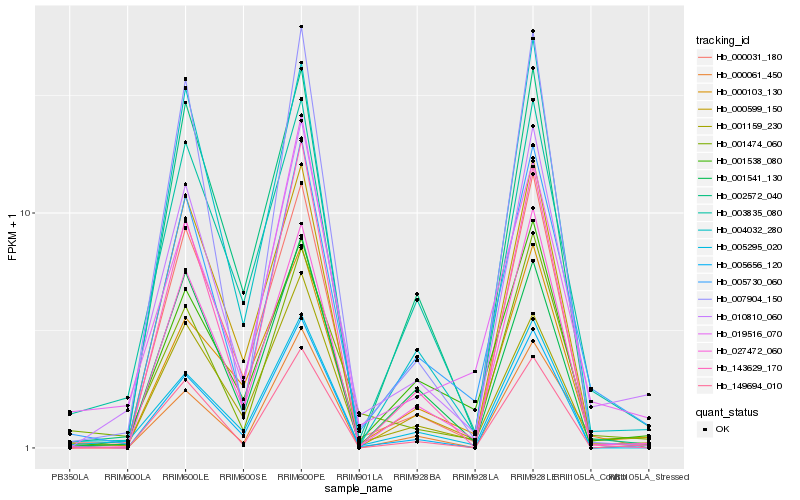
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005730_060 |
0.0 |
- |
- |
PREDICTED: inorganic pyrophosphatase 1-like [Jatropha curcas] |
| 2 |
Hb_010810_060 |
0.0974982447 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 3 |
Hb_027472_060 |
0.1096481717 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 4 |
Hb_003835_080 |
0.1152128789 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001538_080 |
0.1171122297 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001159_230 |
0.1218124605 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 7 |
Hb_000031_180 |
0.1271385141 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 8 |
Hb_000103_130 |
0.1275072276 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 9 |
Hb_019516_070 |
0.1289865932 |
- |
- |
PREDICTED: uncharacterized protein LOC105634258 [Jatropha curcas] |
| 10 |
Hb_001474_060 |
0.1314776845 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 11 |
Hb_000061_450 |
0.1316980959 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001541_130 |
0.1326809978 |
- |
- |
GDSL esterase/lipase [Morus notabilis] |
| 13 |
Hb_004032_280 |
0.1343722254 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 14 |
Hb_002572_040 |
0.1350029512 |
- |
- |
PREDICTED: uncharacterized protein LOC105635149 [Jatropha curcas] |
| 15 |
Hb_005656_120 |
0.1362447772 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 16 |
Hb_149694_010 |
0.1366466707 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 17 |
Hb_000599_150 |
0.1369904267 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 18 |
Hb_143629_170 |
0.1382644835 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 19 |
Hb_005295_020 |
0.1406470932 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Populus euphratica] |
| 20 |
Hb_007904_150 |
0.1408688921 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |