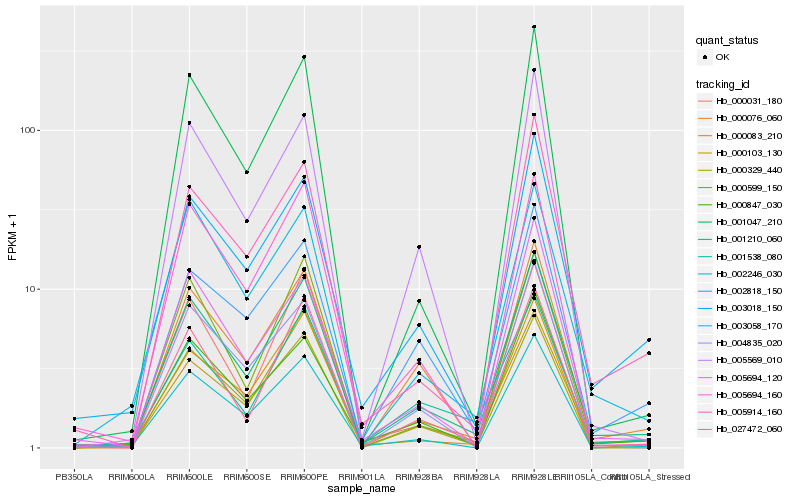
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001210_060 |
0.0 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 2 |
Hb_000847_030 |
0.0818759778 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 3 |
Hb_003018_150 |
0.0856652834 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 4 |
Hb_002246_030 |
0.0890611893 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_000329_440 |
0.091610553 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 6 |
Hb_027472_060 |
0.0946631497 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 7 |
Hb_000103_130 |
0.0948638386 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 8 |
Hb_001047_210 |
0.0977076755 |
- |
- |
PSI reaction center subunit II [Citrus sinensis] |
| 9 |
Hb_002818_150 |
0.0990888779 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 10 |
Hb_000031_180 |
0.0992959482 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 11 |
Hb_005569_010 |
0.1013821454 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000599_150 |
0.1014006515 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 13 |
Hb_000076_060 |
0.101447942 |
- |
- |
hypothetical protein RCOM_0586940 [Ricinus communis] |
| 14 |
Hb_005694_120 |
0.1024147758 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 15 |
Hb_003058_170 |
0.1024193843 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005914_160 |
0.1029461485 |
- |
- |
PREDICTED: protein PROTON GRADIENT REGULATION 5, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001538_080 |
0.1041441038 |
- |
- |
PREDICTED: QWRF motif-containing protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_004835_020 |
0.1052039839 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 19 |
Hb_005694_160 |
0.1057510055 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000083_210 |
0.1063386167 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |