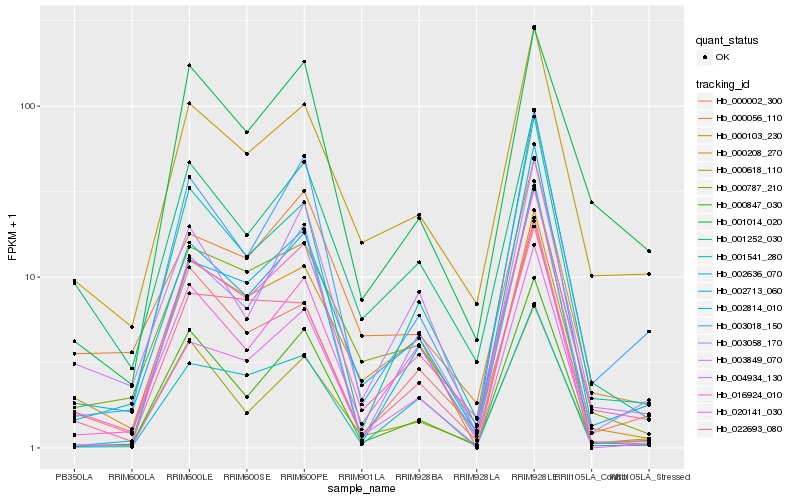
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020141_030 |
0.0 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 2 |
Hb_000208_270 |
0.0850743713 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 3 |
Hb_000787_210 |
0.0933143133 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001541_280 |
0.1004823039 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 5 |
Hb_003018_150 |
0.1025731249 |
- |
- |
PREDICTED: uncharacterized protein LOC105628869 [Jatropha curcas] |
| 6 |
Hb_002814_010 |
0.1081370658 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003058_170 |
0.1106183478 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000002_300 |
0.1129106542 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 9 |
Hb_000847_030 |
0.1133891155 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 10 |
Hb_001252_030 |
0.1149436873 |
- |
- |
Serine/threonine-protein kinase Nek8, putative [Ricinus communis] |
| 11 |
Hb_000103_230 |
0.1168405514 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_016924_010 |
0.1179157269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002636_070 |
0.118987282 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 14 |
Hb_002713_060 |
0.119443541 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 15 |
Hb_004934_130 |
0.1196170388 |
- |
- |
12-oxophytodienoate reductase 2 isoform 1 [Theobroma cacao] |
| 16 |
Hb_000056_110 |
0.1208178072 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 17 |
Hb_001014_020 |
0.1220113281 |
- |
- |
gene GA family protein [Populus trichocarpa] |
| 18 |
Hb_022693_080 |
0.1234115211 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 19 |
Hb_000618_110 |
0.1246743816 |
- |
- |
hypothetical protein POPTR_0003s09870g [Populus trichocarpa] |
| 20 |
Hb_003849_070 |
0.1248141564 |
- |
- |
PREDICTED: uncharacterized protein LOC105644690 [Jatropha curcas] |