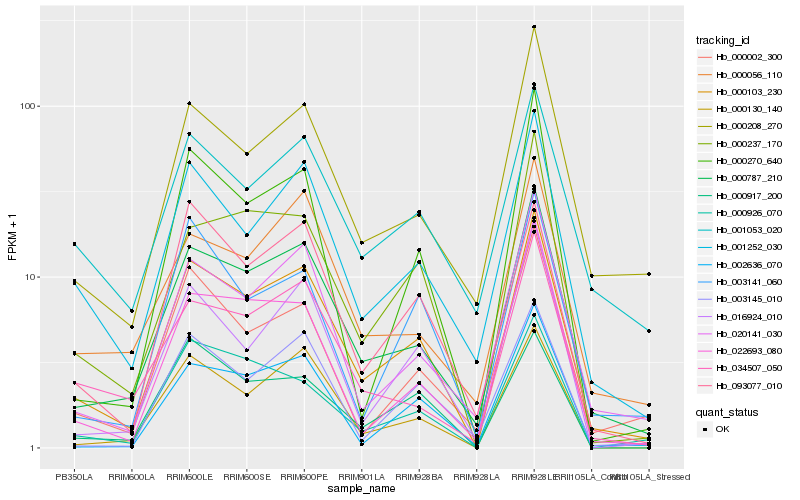
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_230 |
0.0 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000787_210 |
0.081131258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001252_030 |
0.1097463767 |
- |
- |
Serine/threonine-protein kinase Nek8, putative [Ricinus communis] |
| 4 |
Hb_020141_030 |
0.1168405514 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 5 |
Hb_000130_140 |
0.1195849 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000237_170 |
0.1206137455 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 7 |
Hb_000208_270 |
0.1316067972 |
- |
- |
PREDICTED: malate dehydrogenase [NADP], chloroplastic [Jatropha curcas] |
| 8 |
Hb_002636_070 |
0.1323665823 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 9 |
Hb_016924_010 |
0.1327627607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000270_640 |
0.1334923213 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 11 |
Hb_000002_300 |
0.1340822707 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_000926_070 |
0.1348840233 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_022693_080 |
0.1356891469 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |
| 14 |
Hb_034507_050 |
0.1368301379 |
- |
- |
PREDICTED: uncharacterized protein LOC105646633 isoform X2 [Jatropha curcas] |
| 15 |
Hb_093077_010 |
0.1368387038 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000917_200 |
0.1387828936 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 17 |
Hb_003145_010 |
0.1416303368 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g59780 [Jatropha curcas] |
| 18 |
Hb_003141_060 |
0.1428429741 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 19 |
Hb_001053_020 |
0.1436122041 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 20 |
Hb_000056_110 |
0.1439373427 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |