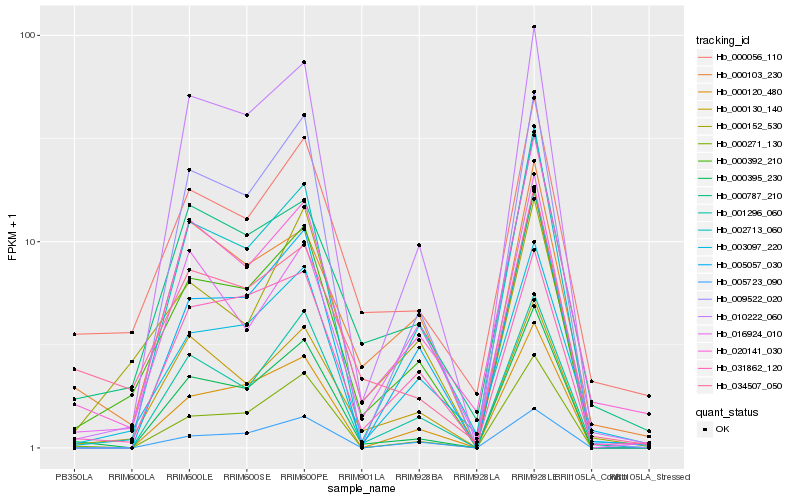
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_210 |
0.0 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |
| 2 |
Hb_000130_140 |
0.1048004816 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000152_530 |
0.1146333267 |
- |
- |
hypothetical protein SELMODRAFT_231485 [Selaginella moellendorffii] |
| 4 |
Hb_010222_060 |
0.1231111626 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_003097_220 |
0.1250627737 |
- |
- |
hypothetical protein POPTR_0001s06150g [Populus trichocarpa] |
| 6 |
Hb_031862_120 |
0.1289033286 |
- |
- |
PREDICTED: root phototropism protein 3 [Jatropha curcas] |
| 7 |
Hb_002713_060 |
0.1296132884 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.4 [Jatropha curcas] |
| 8 |
Hb_000395_230 |
0.1312018926 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Vitis vinifera] |
| 9 |
Hb_000787_210 |
0.1341798436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000056_110 |
0.1376102158 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 11 |
Hb_009522_020 |
0.1404762818 |
- |
- |
PREDICTED: protein ECERIFERUM 3 [Jatropha curcas] |
| 12 |
Hb_034507_050 |
0.1410819642 |
- |
- |
PREDICTED: uncharacterized protein LOC105646633 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001296_060 |
0.1417477468 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_005057_030 |
0.1441058147 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 15 |
Hb_000120_480 |
0.1452900769 |
- |
- |
- |
| 16 |
Hb_005723_090 |
0.1460508197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_016924_010 |
0.1462174327 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000271_130 |
0.148881803 |
transcription factor |
TF Family: C2C2-Dof |
DNA binding with one finger 2.4, putative [Theobroma cacao] |
| 19 |
Hb_020141_030 |
0.1489200727 |
- |
- |
solanesyl diphosphate synthase, putative [Ricinus communis] |
| 20 |
Hb_000103_230 |
0.1489687592 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |