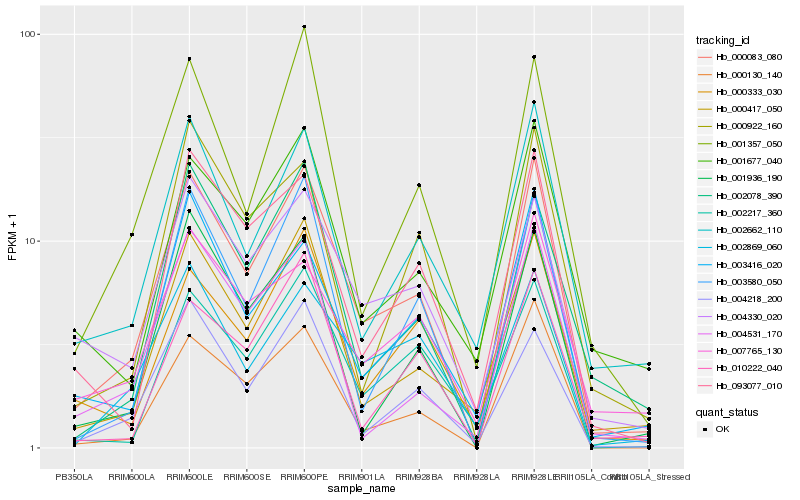
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 2 |
Hb_000130_140 |
0.1086022737 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639203 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003580_050 |
0.1202161028 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 4 |
Hb_000417_050 |
0.1216464653 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase MRH1 [Jatropha curcas] |
| 5 |
Hb_000333_030 |
0.1304834545 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002662_110 |
0.1329586438 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 7 |
Hb_001936_190 |
0.1329751946 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 8 |
Hb_004218_200 |
0.1340794585 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 9 |
Hb_001357_050 |
0.1352617338 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 10 |
Hb_003416_020 |
0.1381523801 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 11 |
Hb_093077_010 |
0.1388654375 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002078_390 |
0.1398008558 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 2 [Jatropha curcas] |
| 13 |
Hb_004330_020 |
0.1402052144 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 14 |
Hb_002217_360 |
0.141262268 |
- |
- |
PREDICTED: nuclear poly(A) polymerase 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002869_060 |
0.1444581858 |
- |
- |
PREDICTED: kinesin-related protein 4 [Jatropha curcas] |
| 16 |
Hb_000922_160 |
0.1449003242 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 17 |
Hb_010222_040 |
0.1459427458 |
- |
- |
PREDICTED: kinesin-2 [Jatropha curcas] |
| 18 |
Hb_007765_130 |
0.1476368387 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 19 |
Hb_004531_170 |
0.1477123869 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 20 |
Hb_001677_040 |
0.1488944448 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 isoform X2 [Jatropha curcas] |