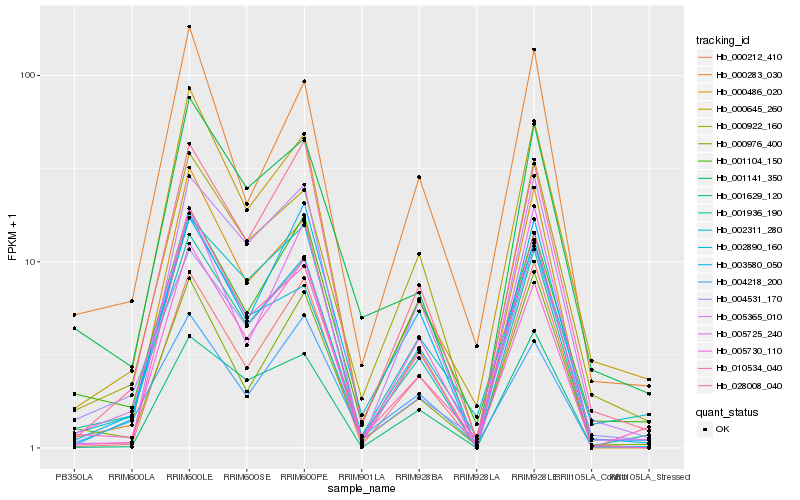
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001936_190 |
0.0 |
- |
- |
hypothetical protein JCGZ_17780 [Jatropha curcas] |
| 2 |
Hb_000922_160 |
0.0930901779 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose 5-phosphate reductoisomerase |
putative 1-deoxy-D-xylulose 5-phosphate reductoisomerase [Hevea brasiliensis] |
| 3 |
Hb_028008_040 |
0.0947858754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_005725_240 |
0.1022116563 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH74 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004531_170 |
0.1033190442 |
- |
- |
PREDICTED: probable inactive nicotinamidase At3g16190 [Jatropha curcas] |
| 6 |
Hb_003580_050 |
0.1073650117 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 7 |
Hb_005365_010 |
0.1094877782 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 8 |
Hb_000486_020 |
0.1102022424 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.2 [Jatropha curcas] |
| 9 |
Hb_005730_110 |
0.110616276 |
- |
- |
PREDICTED: inactive protein kinase SELMODRAFT_444075 [Jatropha curcas] |
| 10 |
Hb_000645_260 |
0.1110560457 |
- |
- |
PREDICTED: uncharacterized protein LOC105643508 [Jatropha curcas] |
| 11 |
Hb_001629_120 |
0.1118122722 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 12 |
Hb_000976_400 |
0.1122834921 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 13 |
Hb_001141_350 |
0.1126097784 |
- |
- |
hypothetical protein JCGZ_02174 [Jatropha curcas] |
| 14 |
Hb_002311_280 |
0.1141857038 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_001104_150 |
0.1161606663 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 16 |
Hb_000212_410 |
0.1168361122 |
- |
- |
PREDICTED: serine carboxypeptidase 1-like [Jatropha curcas] |
| 17 |
Hb_002890_160 |
0.1178451326 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000283_030 |
0.1191117623 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA16-like [Populus euphratica] |
| 19 |
Hb_010534_040 |
0.1206731391 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004218_200 |
0.1230685602 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |