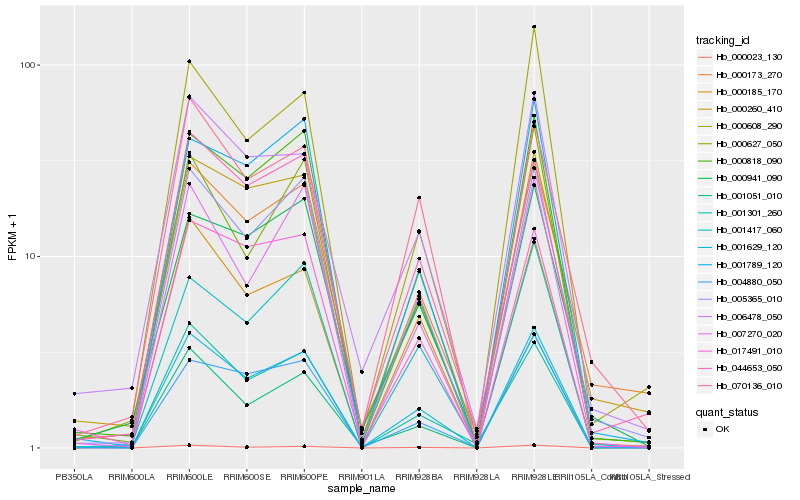
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001629_120 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Populus euphratica] |
| 2 |
Hb_005365_010 |
0.0508319832 |
- |
- |
PREDICTED: serine carboxypeptidase-like 20 [Jatropha curcas] |
| 3 |
Hb_000173_270 |
0.079133649 |
- |
- |
PREDICTED: mitogen-activated protein kinase 15 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000818_090 |
0.0835487921 |
- |
- |
glutamate receptor 3 plant, putative [Ricinus communis] |
| 5 |
Hb_000023_130 |
0.0857701483 |
- |
- |
Metal-nicotianamine transporter YSL1 [Glycine soja] |
| 6 |
Hb_000185_170 |
0.089053074 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_001051_010 |
0.0892451924 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 [Jatropha curcas] |
| 8 |
Hb_001417_060 |
0.0894720005 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 9 |
Hb_000260_410 |
0.093085166 |
- |
- |
PREDICTED: protein LURP-one-related 8 [Jatropha curcas] |
| 10 |
Hb_006478_050 |
0.0945193298 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004880_050 |
0.0964288418 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000627_050 |
0.0970677537 |
- |
- |
PREDICTED: uncharacterized protein LOC105632845 [Jatropha curcas] |
| 13 |
Hb_001301_260 |
0.0979156705 |
- |
- |
PREDICTED: 1-acyl-sn-glycerol-3-phosphate acyltransferase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_044653_050 |
0.098315647 |
- |
- |
PREDICTED: CTL-like protein DDB_G0274487 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000608_290 |
0.0983661067 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 16 |
Hb_001789_120 |
0.099574479 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000941_090 |
0.1013059374 |
- |
- |
PREDICTED: protein trichome birefringence [Jatropha curcas] |
| 18 |
Hb_007270_020 |
0.1021982668 |
- |
- |
hypothetical protein JCGZ_09593 [Jatropha curcas] |
| 19 |
Hb_070136_010 |
0.1024663378 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 20 |
Hb_017491_010 |
0.1027521241 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |